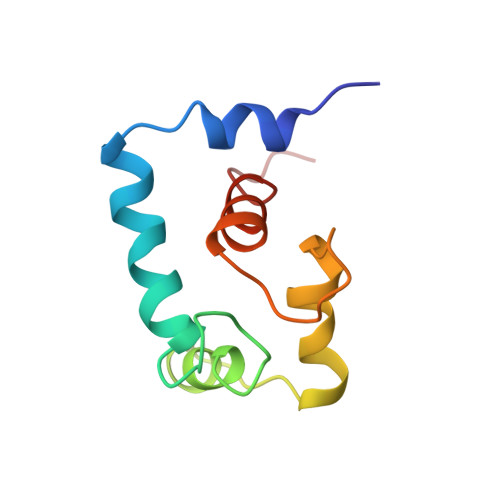
Binding of cardiac troponin-I147-163 induces a structural opening in human cardiac troponin-C.
Li, M.X., Spyracopoulos, L., Sykes, B.D.(1999) Biochemistry 38: 8289-8298
- PubMed: 10387074
- DOI: https://doi.org/10.1021/bi9901679
- Primary Citation of Related Structures:
1MXL - PubMed Abstract:
The interaction of troponin-C (TnC) with troponin-I (TnI) plays a central role in skeletal and cardiac muscle contraction. We have recently shown that the binding of Ca2+ to cardiac TnC (cTnC) does not induce an "opening" of the regulatory domain in order to interact with cTnI [Sia, S. K., et al. (1997) J. Biol. Chem. 272, 18216-18221; Spyracopoulos et al. (1997) Biochemistry 36, 12138-12146], which is in contrast to the regulatory N-domain of skeletal TnC (sTnC). This implies that the mode of interaction between cTnC and cTnI may be different than that between sTnC and sTnI. In sTnI, a region downstream from the inhibitory region (residues 115-131) has been shown to bind the exposed hydrophobic pocket of Ca2+-saturated sNTnC [McKay, R. T., et al. (1997) J. Biol. Chem. 272, 28494-28500]. The present study demonstrates that the corresponding region in cTnI (residues 147-163) binds to the regulatory domain of cTnC only in the Ca2+-saturated state to form a 1:1 complex, with an affinity approximately six times weaker than that between the skeletal counterparts. Thus, while Ca2+ does not cause opening, it is required for muscle regulation. The solution structure of the cNTnC.Ca2+.cTnI147-163 complex has been determined by multinuclear multidimensional NMR spectroscopy. The structure reveals an open conformation for cNTnC, similar to that of Ca2+-saturated sNTnC. The bound peptide adopts a alpha-helical conformation spanning residues 150-157. The C-terminus of the peptide is unstructured. The open conformation for Ca2+-saturated cNTnC in the presence of cTnI (residues 147-163) accommodates hydrophobic interactions between side chains of the peptide and side chains at the interface of A and B helices of cNTnC. Thus the mechanistic differences between the regulation of cardiac and skeletal muscle contraction can be understood in terms of different thermodynamics and kinetics equilibria between essentially the same structure states.
Organizational Affiliation:
MRC Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, Canada.