Crystal structure of a small heat-shock protein.
Kim, K.K., Kim, R., Kim, S.H.(1998) Nature 394: 595-599
- PubMed: 9707123
- DOI: https://doi.org/10.1038/29106
- Primary Citation of Related Structures:
1SHS - PubMed Abstract:
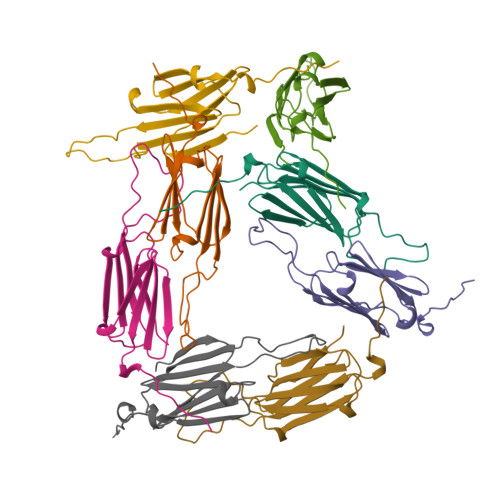
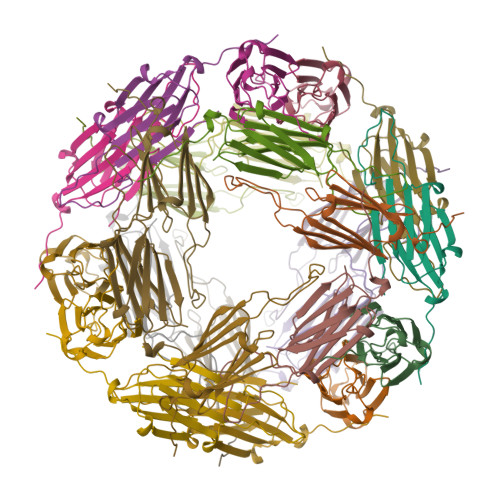
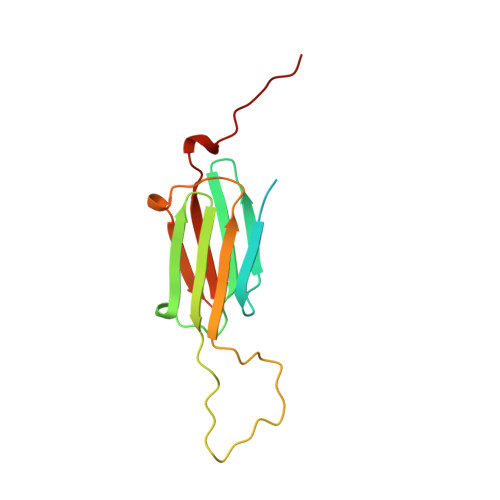
The principal heat-shock proteins that have chaperone activity (that is, they protect newly made proteins from misfolding) belong to five conserved classes: HSP100, HSP90, HSP70, HSP60 and the small heat-shock proteins (sHSPs). The sHSPs can form large multimeric structures and have a wide range of cellular functions, including endowing cells with thermotolerance in vivo and being able to act as molecular chaperones in vitro; sHSPs do this by forming stable complexes with folding intermediates of their protein substrates. However, there is little information available about these structures or the mechanism by which substrates are protected from thermal denaturation by sHSPs. Here we report the crystal structure of a small heat-shock protein from Methanococcus jannaschii, a hyperthermophilic archaeon. The monomeric folding unit is a composite beta-sandwich in which one of the beta-strands comes from a neighbouring molecule. Twenty-four monomers form a hollow spherical complex of octahedral symmetry, with eight trigonal and six square 'windows'. The sphere has an outer diameter of 120 A and an inner diameter of 65 A.
Organizational Affiliation:
Physical Biosciences Division of the Lawrence Berkeley National Laboratory and the Department of Chemistry, University of California at Berkeley, 94720-5230, USA.