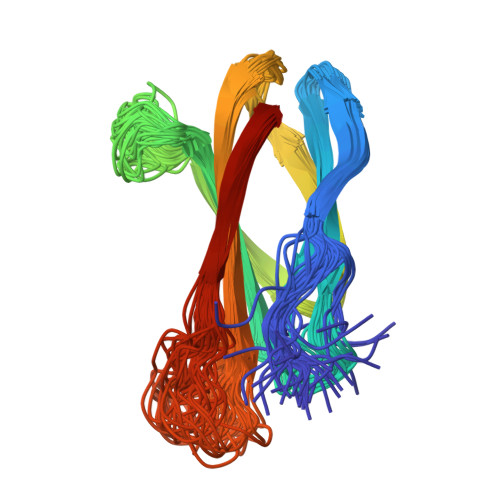
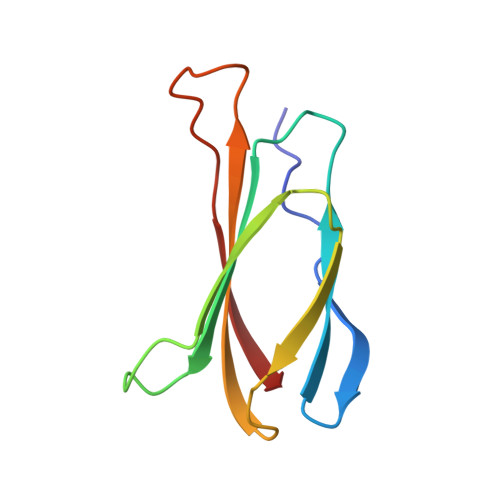
The three-dimensional structure of the tenth type III module of fibronectin: an insight into RGD-mediated interactions.
Main, A.L., Harvey, T.S., Baron, M., Boyd, J., Campbell, I.D.(1992) Cell 71: 671-678
- PubMed: 1423622
- DOI: https://doi.org/10.1016/0092-8674(92)90600-h
- Primary Citation of Related Structures:
1TTF, 1TTG - PubMed Abstract:
The solution structure of the tenth type III module of fibronectin has been determined using nuclear magnetic resonance techniques. The molecule has a fold similar to that of immunoglobulin domains, with seven beta strands forming two antiparallel beta sheets, which pack against each other. Both beta sheets contribute conserved hydrophobic residues to a compact core. The topology is more similar to that of domain 2 of CD4, PapD, and the extracellular domain of the human growth hormone receptor than to that of immunoglobulin C domains. The module contains an Arg-Gly-Asp sequence known to be involved in cell adhesion. This tripeptide is solvent exposed and lies on a conformationally mobile loop between strands F and G, consistent with its cell adhesion function.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, England.