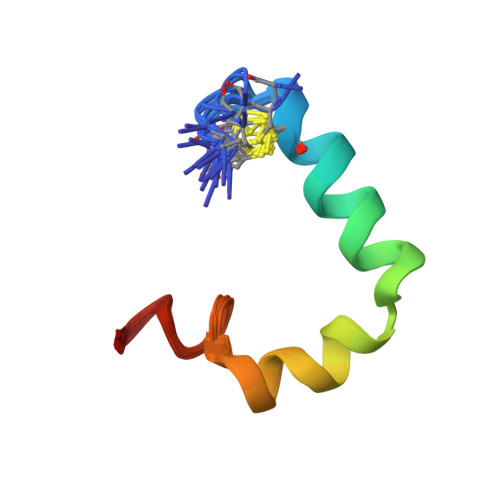
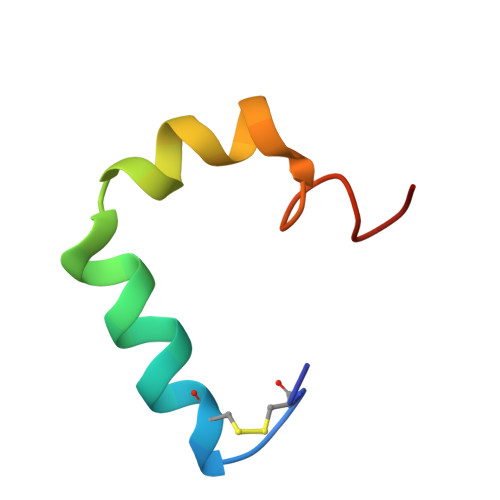
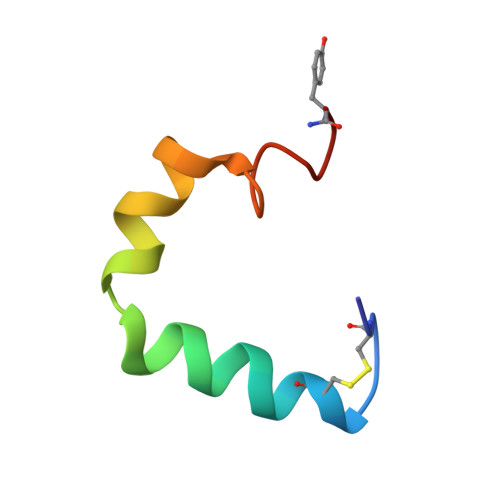
Structure and membrane orientation of IAPP in its natively amidated form at physiological pH in a membrane environment.
Nanga, R.P., Brender, J.R., Vivekanandan, S., Ramamoorthy, A.(2011) Biochim Biophys Acta 1808: 2337-2342
- PubMed: 21723249
- DOI: https://doi.org/10.1016/j.bbamem.2011.06.012
- Primary Citation of Related Structures:
2L86 - PubMed Abstract:
Human islet amyloid polypeptide is a hormone coexpressed with insulin by pancreatic beta-cells. For reasons not clearly understood, hIAPP aggregates in type II diabetics to form oligomers that interfere with beta-cell function, eventually leading to the loss of insulin production. The cellular membrane catalyzes the formation of amyloid deposits and is a target of amyloid toxicity through disruption of the membrane's structural integrity. Therefore, there is considerable current interest in solving the 3D structure of this peptide in a membrane environment. NMR experiments could not be directly utilized in lipid bilayers due to the rapid aggregation of the peptide. To overcome this difficulty, we have solved the structure of the naturally occurring peptide in detergent micelles at a neutral pH. The structure has an overall kinked helix motif, with residues 7-17 and 21-28 in a helical conformation, and with a 3(10) helix from Gly 33-Asn 35. In addition, the angle between the N- and C-terminal helices is constrained to 85°. The greater helical content of human IAPP in the amidated versus free acid form is likely to play a role in its aggregation and membrane disruptive activity.
Organizational Affiliation:
Department of Chemistry, University of Michigan, Ann Arbor, MI 48109-1055, USA.