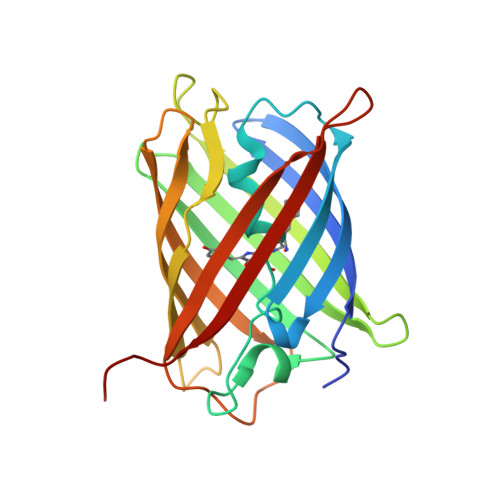
Three-dimensional structure of yellow fluorescent protein zYFP538 from Zoanthus sp. at the resolution 1.8 angstrom
Pletneva, N.V., Pletnev, S.V., Chudakov, D.M., Tikhonova, T.V., Popov, V.O., Martynov, V.I., Wlodawer, A., Dauter, Z., Pletnev, V.Z.(null) Bioorg Khim 33: 421-430
- PubMed: 17886433
- DOI: https://doi.org/10.1134/s1068162007040048
- Primary Citation of Related Structures:
2OGR - PubMed Abstract:
The three-dimensional structure of yellow fluorescent proteins zYFP538 (zFP538) from the button polyp Zoanthus sp. was determined at a resolution of 1.8 angstrom by X-ray analysis. The monomer of zYFP538 adopts a structure characteristic of the green fluorescent protein (GFP) family, a beta-barrel formed from 11 antiparallel beta segments and one internal alpha helix with a chromophore embedded into it. Like the TurboGFP, the beta-barrel of zYFP538 contains a water-filled pore leading to the chromophore Tyr67 residue, which presumably provides access of molecular oxygen necessary for the maturation process. The post-translational modification of the chromophore-forming triad Lys66-Tyr67-Gly68 results in a tricyclic structure consisting of a five-membered imidazolinone ring, a phenol ring of the Tyr67 residue, and an additional six-membered tetrahydropyridine ring. The chromophore formation is completed by cleavage of the protein backbone at the Calpha-N bond of Lys66. It was suggested that the energy conflict between the buried positive charge of the intact Lys66 side chain in the hydrophobic pocket formed by the Ile44, Leu46, Phe65, Leu204 and Leu219 side chains is the most probable trigger that induces the transformation of the bicyclic green form to the tricyclic yellow form. A stereochemical analysis of the contacting surfaces at the intratetramer interfaces helped reveal a group of conserved key residues responsible for the oligomerization. Along with others, these residues should be taken into account in designing monomeric forms suitable for practical application as markers of proteins and cell organelles.