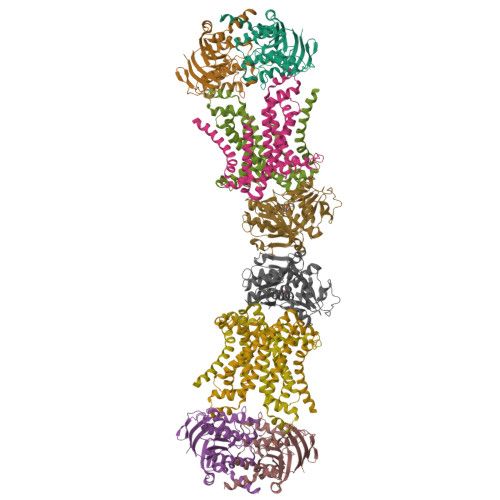
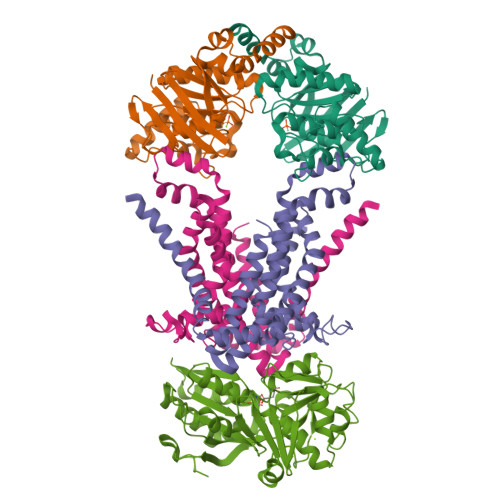
Structure of an ABC transporter in complex with its binding protein.
Hollenstein, K., Frei, D.C., Locher, K.P.(2007) Nature 446: 213-216
- PubMed: 17322901
- DOI: https://doi.org/10.1038/nature05626
- Primary Citation of Related Structures:
2ONK, 2ONR, 2ONS - PubMed Abstract:
ATP-binding cassette (ABC) transporter proteins carry diverse substrates across cell membranes. Whereas clinically relevant ABC exporters are implicated in various diseases or cause multidrug resistance of cancer cells, bacterial ABC importers are essential for the uptake of nutrients, including rare elements such as molybdenum. A detailed understanding of their mechanisms requires direct visualization at high resolution and in distinct conformations. Our recent structure of the multidrug ABC exporter Sav1866 has revealed an outward-facing conformation of the transmembrane domains coupled to a closed conformation of the nucleotide-binding domains, reflecting the ATP-bound state. Here we present the 3.1 A crystal structure of a putative molybdate transporter (ModB2C2) from Archaeoglobus fulgidus in complex with its binding protein (ModA). Twelve transmembrane helices of the ModB subunits provide an inward-facing conformation, with a closed gate near the external membrane boundary. The ATP-hydrolysing ModC subunits reveal a nucleotide-free, open conformation, whereas the attached binding protein aligns the substrate-binding cleft with the entrance to the presumed translocation pathway. Structural comparison of ModB2C2A with Sav1866 suggests a common alternating access and release mechanism, with binding of ATP promoting an outward-facing conformation and dissociation of the hydrolysis products promoting an inward-facing conformation.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zurich, 8093 Zurich, Switzerland.