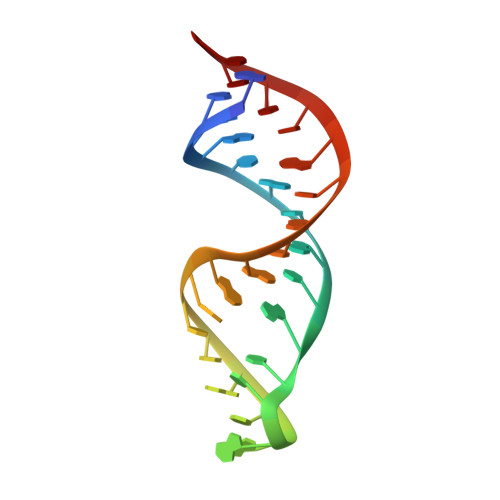
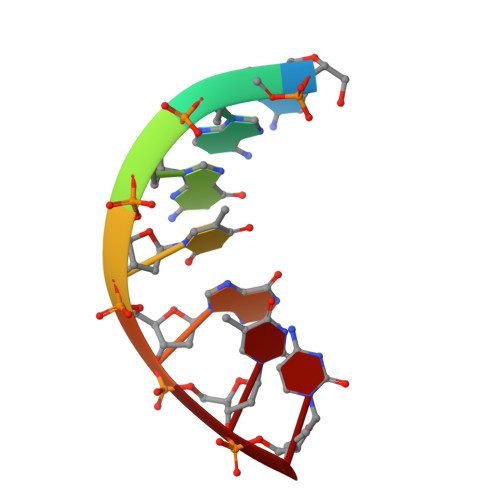
The Role of Mg(II) in DNA Cleavage Site Recognition in Group II Intron Ribozymes: SOLUTION STRUCTURE AND METAL ION BINDING SITES OF THE RNADNA COMPLEX.
Skilandat, M., Sigel, R.K.(2014) J Biol Chem 289: 20650-20663
- PubMed: 24895129
- DOI: https://doi.org/10.1074/jbc.M113.542381
- Primary Citation of Related Structures:
2M1V - PubMed Abstract:
Group II intron ribozymes catalyze the cleavage of (and their reinsertion into) DNA and RNA targets using a Mg2(+)-dependent reaction. The target is cleaved 3' to the last nucleotide of intron binding site 1 (IBS1), one of three regions that form base pairs with the intron's exon binding sites (EBS1 to -3).We solved the NMR solution structure of the d3' hairpin of the Sc.ai5γ intron containing EBS1 in its 11-nucleotide loop in complex with the dIBS1 DNA 7-mer and compare it with the analogous RNA-RNA contact. The EBS1-dIBS1 helix is slightly flexible and non-symmetric. NMR data reveal two major groove binding sites for divalent metal ions at the EBS1-dIBS1 helix, and surface plasmon resonance experiments show that low concentrations of Mg2(+) considerably enhance the affinity of dIBS1 for EBS1. Our results indicate that identification of both RNA and DNA IBS1 targets, presentation of the scissile bond, and stabilization of the structure by metal ions are governed by the overall structure of EBS1-dIBS1 and the surrounding loop nucleotides but are irrespective of different EBS1-(d)IBS1 geometries and interstrand affinities.