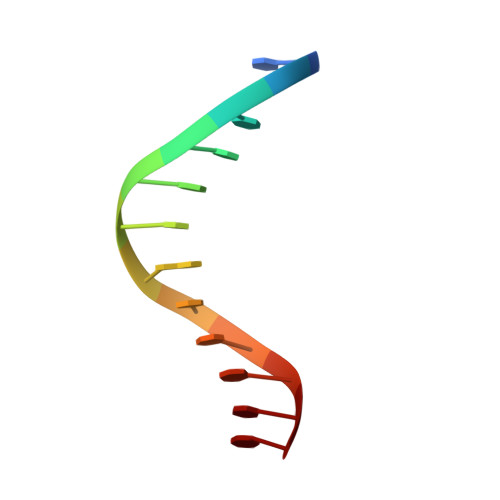
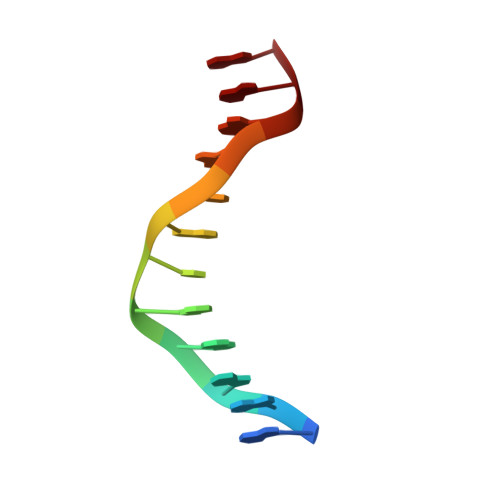
cis-Diammine(pyridine)chloroplatinum(II), a monofunctional platinum(II) antitumor agent: Uptake, structure, function, and prospects.
Lovejoy, K.S., Todd, R.C., Zhang, S., McCormick, M.S., D'Aquino, J.A., Reardon, J.T., Sancar, A., Giacomini, K.M., Lippard, S.J.(2008) Proc Natl Acad Sci U S A 105: 8902-8907
- PubMed: 18579768
- DOI: https://doi.org/10.1073/pnas.0803441105
- Primary Citation of Related Structures:
3CO3 - PubMed Abstract:
We have identified unique chemical and biological properties of a cationic monofunctional platinum(II) complex, cis-diammine(pyridine)chloroplatinum(II), cis-[Pt(NH(3))(2)(py)Cl](+) or cDPCP, a coordination compound previously identified to have significant anticancer activity in a mouse tumor model. This compound is an excellent substrate for organic cation transporters 1 and 2, also designated SLC22A1 and SLC22A2, respectively. These transporters are abundantly expressed in human colorectal cancers, where they mediate uptake of oxaliplatin, cis-[Pt(DACH)(oxalate)] (DACH = trans-R,R-1,2-diaminocyclohexane), an FDA-approved first-line therapy for colorectal cancer. Unlike oxaliplatin, however, cDPCP binds DNA monofunctionally, as revealed by an x-ray crystal structure of cis-{Pt(NH(3))(2)(py)}(2+) bound to the N7 atom of a single guanosine residue in a DNA dodecamer duplex. Although the quaternary structure resembles that of B-form DNA, there is a base-pair step to the 5' side of the Pt adduct with abnormally large shift and slide values, features characteristic of cisplatin intrastrand cross-links. cDPCP effectively blocks transcription from DNA templates carrying adducts of the complex, unlike DNA lesions of other monofunctional platinum(II) compounds like {Pt(dien)}(2+). cDPCP-DNA adducts are removed by the nucleotide excision repair apparatus, albeit much less efficiently than bifunctional platinum-DNA intrastrand cross-links. These exceptional characteristics indicate that cDPCP and related complexes merit consideration as therapeutic options for treating colorectal and other cancers bearing appropriate cation transporters.
Organizational Affiliation:
Department of Chemistry, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.