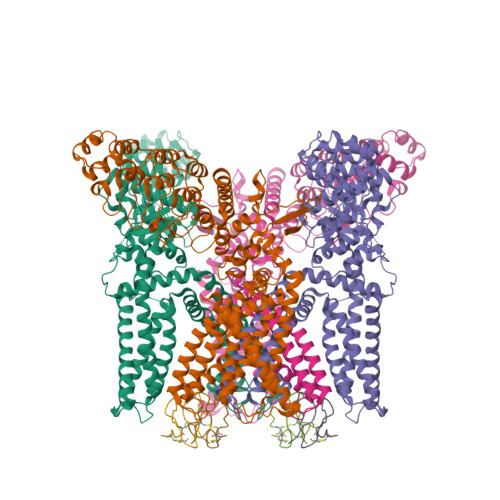
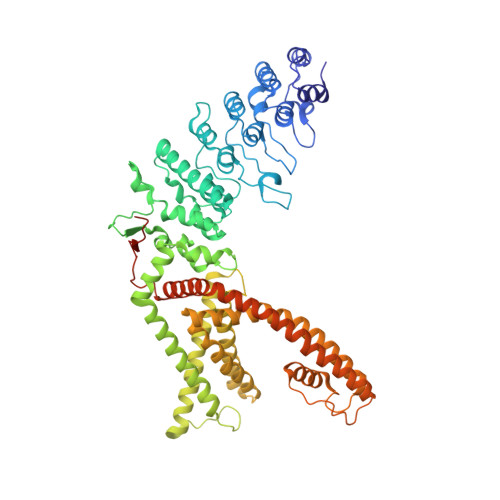
TRPV1 structures in distinct conformations reveal activation mechanisms.
Cao, E., Liao, M., Cheng, Y., Julius, D.(2013) Nature 504: 113-118
- PubMed: 24305161
- DOI: https://doi.org/10.1038/nature12823
- Primary Citation of Related Structures:
3J5Q, 3J5R - PubMed Abstract:
Transient receptor potential (TRP) channels are polymodal signal detectors that respond to a wide range of physical and chemical stimuli. Elucidating how these channels integrate and convert physiological signals into channel opening is essential to understanding how they regulate cell excitability under normal and pathophysiological conditions. Here we exploit pharmacological probes (a peptide toxin and small vanilloid agonists) to determine structures of two activated states of the capsaicin receptor, TRPV1. A domain (consisting of transmembrane segments 1-4) that moves during activation of voltage-gated channels remains stationary in TRPV1, highlighting differences in gating mechanisms for these structurally related channel superfamilies. TRPV1 opening is associated with major structural rearrangements in the outer pore, including the pore helix and selectivity filter, as well as pronounced dilation of a hydrophobic constriction at the lower gate, suggesting a dual gating mechanism. Allosteric coupling between upper and lower gates may account for rich physiological modulation exhibited by TRPV1 and other TRP channels.
Organizational Affiliation:
1] Department of Physiology, University of California, San Francisco, California 94158-2517, USA [2].