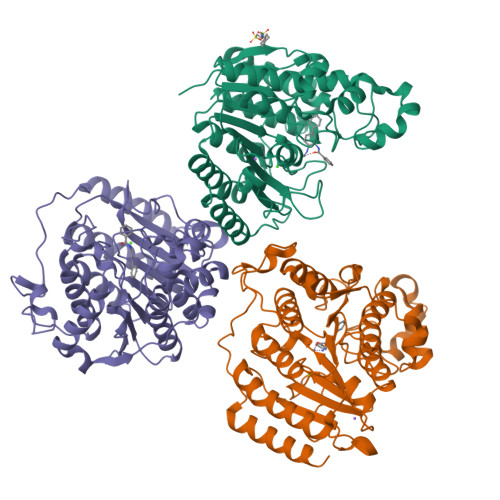
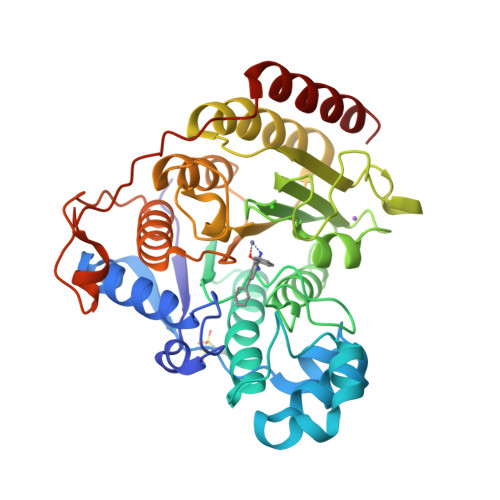
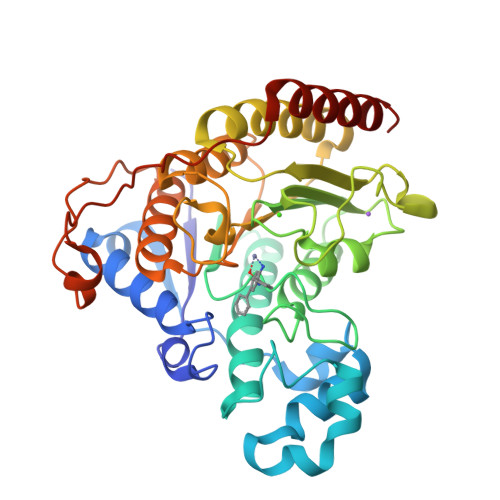
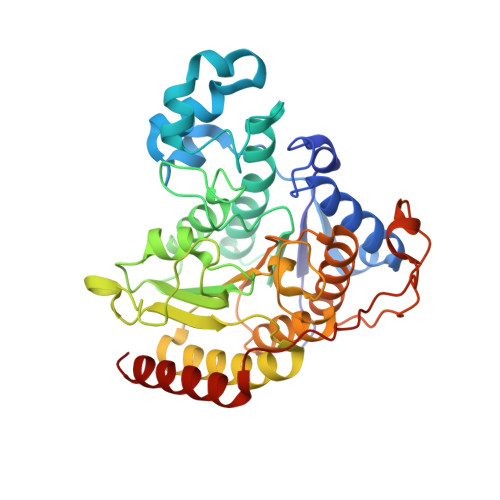
Exploration of the HDAC2 foot pocket: Synthesis and SAR of substituted N-(2-aminophenyl)benzamides.
Bressi, J.C., Jennings, A.J., Skene, R., Wu, Y., Melkus, R., De Jong, R., O'Connell, S., Grimshaw, C.E., Navre, M., Gangloff, A.R.(2010) Bioorg Med Chem Lett 20: 3142-3145
- PubMed: 20392638
- DOI: https://doi.org/10.1016/j.bmcl.2010.03.091
- Primary Citation of Related Structures:
3MAX - PubMed Abstract:
A series of N-(2-amino-5-substituted phenyl)benzamides (3-21) were designed, synthesized and evaluated for their inhibition of HDAC2 and their cytotoxicity in HCT116 cancer cells. Multiple compounds from this series demonstrated time-dependent binding kinetics that is rationalized using a co-complex crystal structure of HDAC2 and N-(4-aminobiphenyl-3-yl)benzamide (6).
Organizational Affiliation:
Takeda San Diego, San Diego, CA 92121, USA.