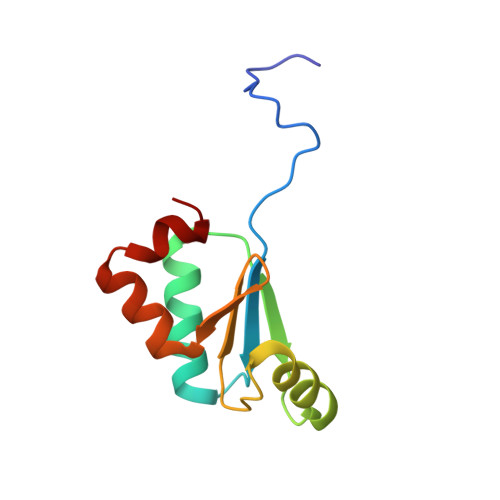
Redox, mutagenic and structural studies of the glutaredoxin/arsenate reductase couple from the cyanobacterium Synechocystis sp. PCC 6803.
Kim, S.G., Chung, J.S., Sutton, R.B., Lee, J.S., Lopez-Maury, L., Lee, S.Y., Florencio, F.J., Lin, T., Zabet-Moghaddam, M., Wood, M.J., Nayak, K., Madem, V., Tripathy, J.N., Kim, S.K., Knaff, D.B.(2011) Biochim Biophys Acta 1824: 392-403
- PubMed: 22155275
- DOI: https://doi.org/10.1016/j.bbapap.2011.10.012
- Primary Citation of Related Structures:
3QMX - PubMed Abstract:
The arsenate reductase from the cyanobacterium Synechocystis sp. PCC 6803 has been characterized in terms of the redox properties of its cysteine residues and their role in the reaction catalyzed by the enzyme. Of the five cysteines present in the enzyme, two (Cys13 and Cys35) have been shown not to be required for catalysis, while Cys8, Cys80 and Cys82 have been shown to be essential. The as-isolated enzyme contains a single disulfide, formed between Cys80 and Cys82, with an oxidation-reduction midpoint potential (E(m)) value of -165mV at pH 7.0. It has been shown that Cys15 is the only one of the four cysteines present in Synechocystis sp. PCC 6803 glutaredoxin A required for its ability to serve as an electron donor to arsenate reductase, while the other three cysteines (Cys18, Cys36 and Cys70) play no role. Glutaredoxin A has been shown to contain a single redox-active disulfide/dithiol couple, with a two-electron, E(m) value of -220mV at pH 7.0. One cysteine in this disulfide/dithiol couple has been shown to undergo glutathionylation. An X-ray crystal structure, at 1.8Å resolution, has been obtained for glutaredoxin A. The probable orientations of arsenate reductase disulfide bonds present in the resting enzyme and in a likely reaction intermediate of the enzyme have been examined by in silico modeling, as has the surface environment of arsenate reductase in the vicinity of Cys8, the likely site for the initial reaction between arsenate and the enzyme.
Organizational Affiliation:
Department of Chemistry and Biochemistry and the Institute of Biomedical Studies, Baylor University, Waco, TX 76798-7348, USA.