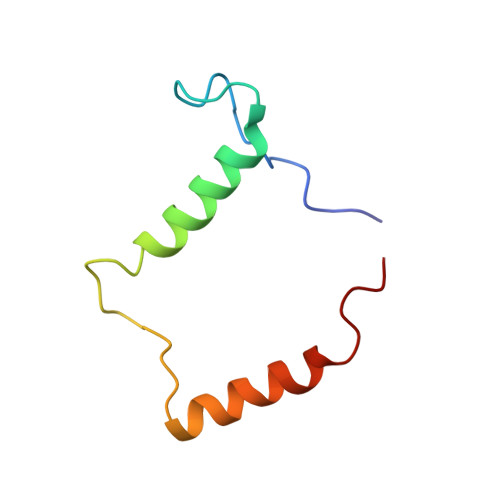
Small-Angle X-Ray Scattering- and Nuclear Magnetic Resonance-Derived Conformational Ensemble of the Highly Flexible Antitoxin Paaa2.
Sterckx, Y.G., Volkov, A.N., Vranken, W.F., Kragelj, J., Jensen, M.R., Buts, L., Garcia-Pino, A., Jove, T., Van Melderen, L., Blackledge, M., Van Nuland, N.A., Loris, R.(2014) Structure 22: 854
- PubMed: 24768114
- DOI: https://doi.org/10.1016/j.str.2014.03.012
- Primary Citation of Related Structures:
3ZBE - PubMed Abstract:
Antitoxins from prokaryotic type II toxin-antitoxin modules are characterized by a high degree of intrinsic disorder. The description of such highly flexible proteins is challenging because they cannot be represented by a single structure. Here, we present a combination of SAXS and NMR data to describe the conformational ensemble of the PaaA2 antitoxin from the human pathogen E. coli O157. The method encompasses the use of SAXS data to filter ensembles out of a pool of conformers generated by a custom NMR structure calculation protocol and the subsequent refinement by a block jackknife procedure. The final ensemble obtained through the method is validated by an established residual dipolar coupling analysis. We show that the conformational ensemble of PaaA2 is highly compact and that the protein exists in solution as two preformed helices, connected by a flexible linker, that probably act as molecular recognition elements for toxin inhibition.
Organizational Affiliation:
Structural Biology Brussels, Department of Biotechnology, Vrije Universiteit Brussel, Pleinlaan 2, B-1050 Brussels, Belgium; Molecular Recognition Unit and Jean Jeener NMR Centre, Structural Biology Research Center, VIB, Pleinlaan 2, B-1050 Brussels, Belgium.