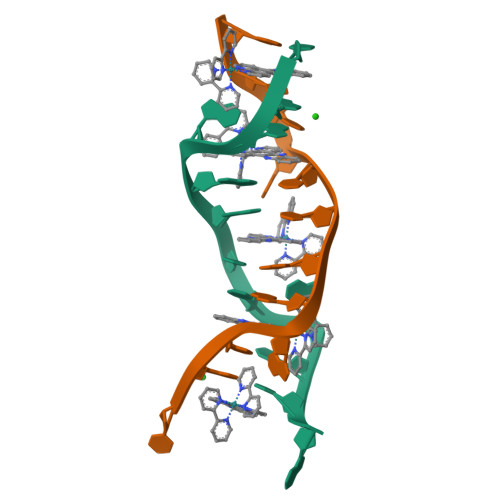
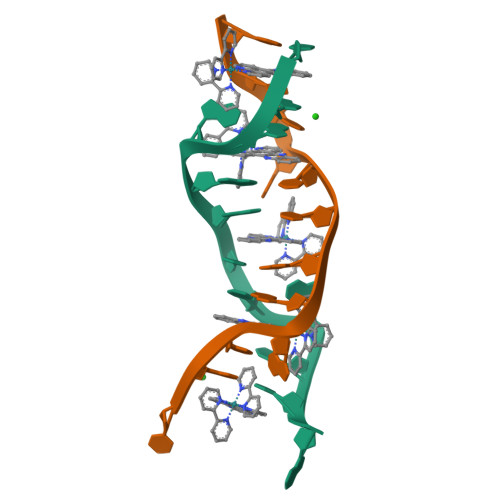
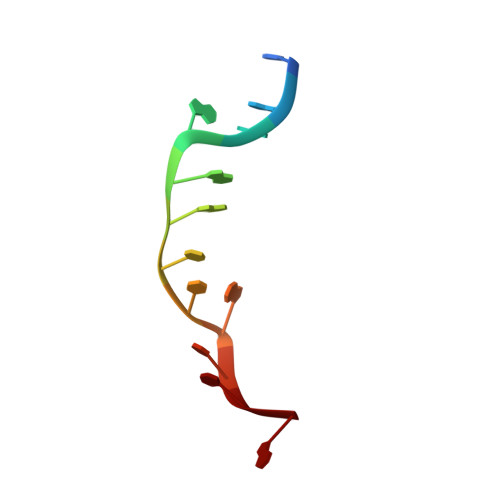
Crystal structure of delta-[Ru(bpy)2dppz]2+ bound to mismatched DNA reveals side-by-side metalloinsertion and intercalation.
Song, H., Kaiser, J.T., Barton, J.K.(2012) Nat Chem 4: 615-620
- PubMed: 22824892
- DOI: https://doi.org/10.1038/nchem.1375
- Primary Citation of Related Structures:
4E1U - PubMed Abstract:
DNA mismatches represent a novel target in the development of diagnostics and therapeutics for cancer, because deficiencies in DNA mismatch repair are implicated in cancers, and cells that are repair-deficient show a high frequency of mismatches. Metal complexes with bulky intercalating ligands serve as probes for DNA mismatches. Here, we report the high-resolution (0.92 Å) crystal structure of the ruthenium 'light switch' complex Δ-[Ru(bpy)(2)dppz](2+) (bpy = 2,2'-bipyridine and dppz = dipyridophenazine), which is known to show luminescence on binding to duplex DNA, bound to both mismatched and well-matched sites in the oligonucleotide 5'-(dCGGAAATTACCG)(2)-3' (underline denotes AA mismatches). Two crystallographically independent views reveal that the complex binds mismatches through metalloinsertion, ejecting both mispaired adenosines. Additional ruthenium complexes are intercalated at well-matched sites, creating an array of complexes in the minor groove stabilized by stacking interactions between bpy ligands and extruded adenosines. This structure attests to the generality of metalloinsertion and metallointercalation as DNA binding modes.
Organizational Affiliation:
Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, California 91125, USA.