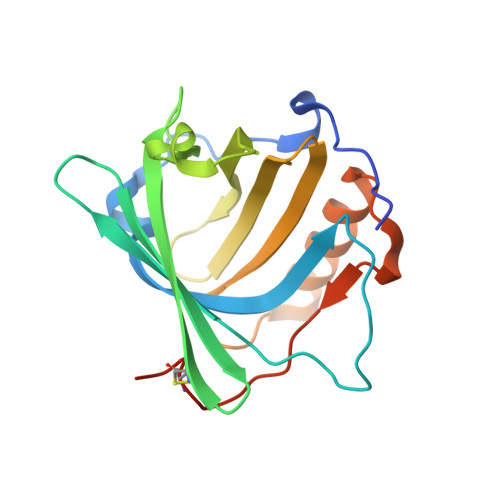
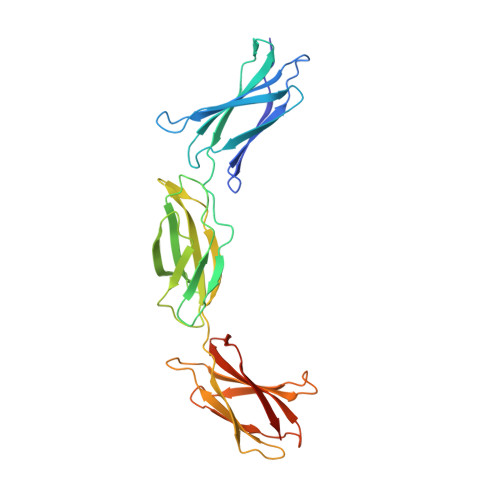
Combinatorial design of an Anticalin directed against the extra-domain b for the specific targeting of oncofetal fibronectin
Gebauer, M., Schiefner, A., Matschiner, G., Skerra, A.(2013) J Mol Biol 425: 780-802
- PubMed: 23238252
- DOI: https://doi.org/10.1016/j.jmb.2012.12.004
- Primary Citation of Related Structures:
4GH7 - PubMed Abstract:
The oncofetal isoform of the extracellular matrix protein fibronectin (Fn), which carries the extra-domain B (ED-B) and is exclusively expressed in neovasculature, has gained interest for tumor diagnosis and therapy using engineered antibody fragments. We have employed the human lipocalin 2 (Lcn2) as a small and robust non-immunoglobulin scaffold to select ED-B-specific Anticalins from a new advanced random library using bacterial phage display and ELISA screening against appropriately engineered Fn fragments. As a result, we have isolated and biochemically characterized four different Anticalins that all show low nanomolar affinities for ED-B, right in the range between the monomeric and dimeric forms of the single-chain variable antibody fragment L19 that has been widely applied in this area before. All Anticalins can be readily expressed in Escherichia coli as soluble and strictly monomeric proteins, and they show specific staining of ED-B-positive tumor cells in immunofluorescence microscopy while BIAcore affinity analyses indicate recognition of distinct ED-B epitopes. The crystal structure for one Anticalin, N7A, in complex with the Fn7B8 fragment, was solved at 2.6Å resolution and reveals binding to the gfcc' sheet and cc' loop on ED-B. This is the second example of a protein-specific Lcn2-based Anticalin, which illustrates the remarkable plasticity of the calyx-like ligand pocket of lipocalins with their four structurally hypervariable loops supported by a highly conserved β-barrel. The ED-B-specific Anticalins resulting from this study should provide useful reagents in research and biomedical drug development, both for in vivo imaging and for directed cancer therapy.
Organizational Affiliation:
Munich Center for Integrated Protein Science (CIPS-M) and Lehrstuhl für Biologische Chemie, Technische Universität München, 85350 Freising-Weihenstephan, Germany.