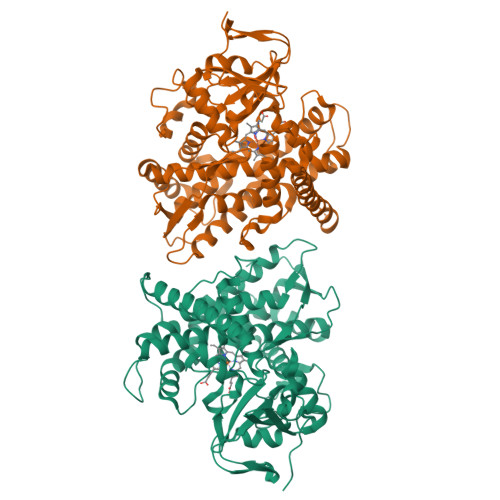
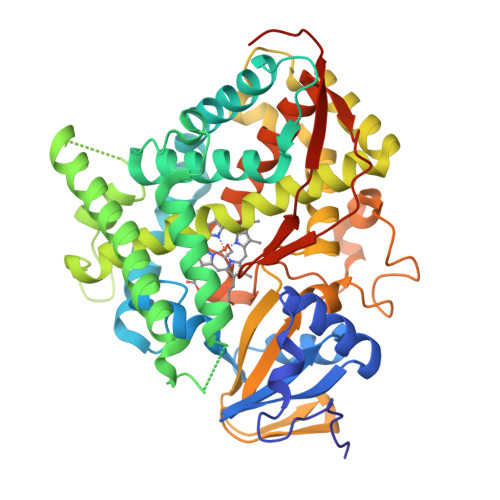
A serine-substituted P450 catalyzes highly efficient carbene transfer to olefins in vivo.
Coelho, P.S., Wang, Z.J., Ener, M.E., Baril, S.A., Kannan, A., Arnold, F.H., Brustad, E.M.(2013) Nat Chem Biol 9: 485-487
- PubMed: 23792734
- DOI: https://doi.org/10.1038/nchembio.1278
- Primary Citation of Related Structures:
4H23, 4H24 - PubMed Abstract:
Whole-cell catalysts for non-natural chemical reactions will open new routes to sustainable production of chemicals. We designed a cytochrome 'P411' with unique serine-heme ligation that catalyzes efficient and selective olefin cyclopropanation in intact Escherichia coli cells. The mutation C400S in cytochrome P450(BM3) gives a signature ferrous CO Soret peak at 411 nm, abolishes monooxygenation activity, raises the resting-state Fe(III)-to-Fe(II) reduction potential and substantially improves NAD(P)H-driven activity.
Organizational Affiliation:
Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena, California, USA.