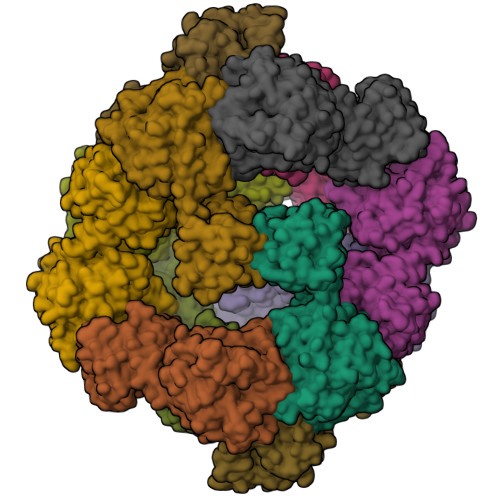
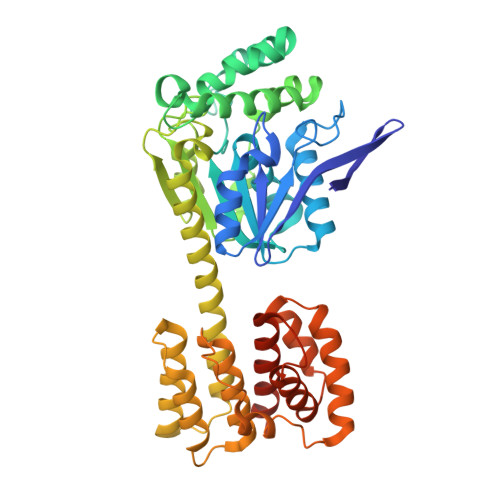
Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly.
Lai, Y.T., Tsai, K.L., Sawaya, M.R., Asturias, F.J., Yeates, T.O.(2013) J Am Chem Soc 135: 7738-7743
- PubMed: 23621606
- DOI: https://doi.org/10.1021/ja402277f
- Primary Citation of Related Structures:
4IQ4, 4ITV, 4IVJ - PubMed Abstract:
Designing protein molecules that self-assemble into complex architectures is an outstanding goal in the area of nanobiotechnology. One design strategy for doing this involves genetically fusing together two natural proteins, each of which is known to form a simple oligomer on its own (e.g., a dimer or trimer). If two such components can be fused in a geometrically predefined configuration, that designed subunit can, in principle, assemble into highly symmetric architectures. Initial experiments showed that a 12-subunit tetrahedral cage, 16 nm in diameter, could be constructed following such a procedure [Padilla, J. E.; et al. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 2217; Lai, Y. T.; et al. Science 2012, 336, 1129]. Here we characterize multiple crystal structures of protein cages constructed in this way, including cages assembled from two mutant forms of the same basic protein subunit. The flexibilities of the designed assemblies and their deviations from the target model are described, along with implications for further design developments.
Organizational Affiliation:
Department of Bioengineering, University of California, Los Angeles, California 90095, USA.