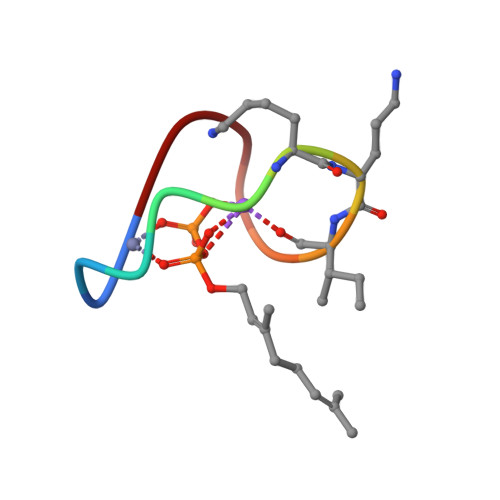
High-resolution crystal structure reveals molecular details of target recognition by bacitracin.
Economou, N.J., Cocklin, S., Loll, P.J.(2013) Proc Natl Acad Sci U S A 110: 14207-14212
- PubMed: 23940351
- DOI: https://doi.org/10.1073/pnas.1308268110
- Primary Citation of Related Structures:
4K7T - PubMed Abstract:
Bacitracin is a metalloantibiotic agent that is widely used as a medicine and feed additive. It interferes with bacterial cell-wall biosynthesis by binding undecaprenyl-pyrophosphate, a lipid carrier that serves as a critical intermediate in cell wall production. Despite bacitracin's broad use, the molecular details of its target recognition have not been elucidated. Here we report a crystal structure for the ternary complex of bacitracin A, zinc, and a geranyl-pyrophosphate ligand at a resolution of 1.1 Å. The antibiotic forms a compact structure that completely envelopes the ligand's pyrophosphate group, together with flanking zinc and sodium ions. The complex adopts a highly amphipathic conformation that offers clues to antibiotic function in the context of bacterial membranes. Bacitracin's efficient sequestration of its target represents a previously unseen mode for the recognition of lipid pyrophosphates, and suggests new directions for the design of next-generation antimicrobial agents.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Drexel University College of Medicine, Philadelphia, PA 19102, USA.