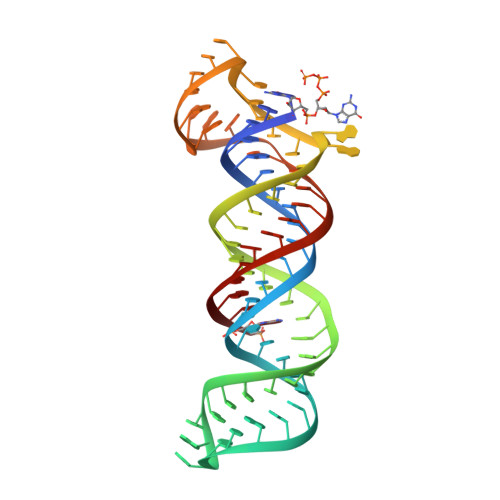
Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix.
Brown, J.A., Bulkley, D., Wang, J., Valenstein, M.L., Yario, T.A., Steitz, T.A., Steitz, J.A.(2014) Nat Struct Mol Biol 21: 633-640
- PubMed: 24952594
- DOI: https://doi.org/10.1038/nsmb.2844
- Primary Citation of Related Structures:
4PLX - PubMed Abstract:
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is a highly abundant nuclear long noncoding RNA that promotes malignancy. A 3'-stem-loop structure is predicted to confer stability by engaging a downstream A-rich tract in a triple helix, similar to the expression and nuclear retention element (ENE) from the KSHV polyadenylated nuclear RNA. The 3.1-Å-resolution crystal structure of the human MALAT1 ENE and A-rich tract reveals a bipartite triple helix containing stacks of five and four U•A-U triples separated by a C+•G-C triplet and C-G doublet, extended by two A-minor interactions. In vivo decay assays indicate that this blunt-ended triple helix, with the 3' nucleotide in a U•A-U triple, inhibits rapid nuclear RNA decay. Interruption of the triple helix by the C-G doublet induces a 'helical reset' that explains why triple-helical stacks longer than six do not occur in nature.
Organizational Affiliation:
Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, Connecticut, USA.