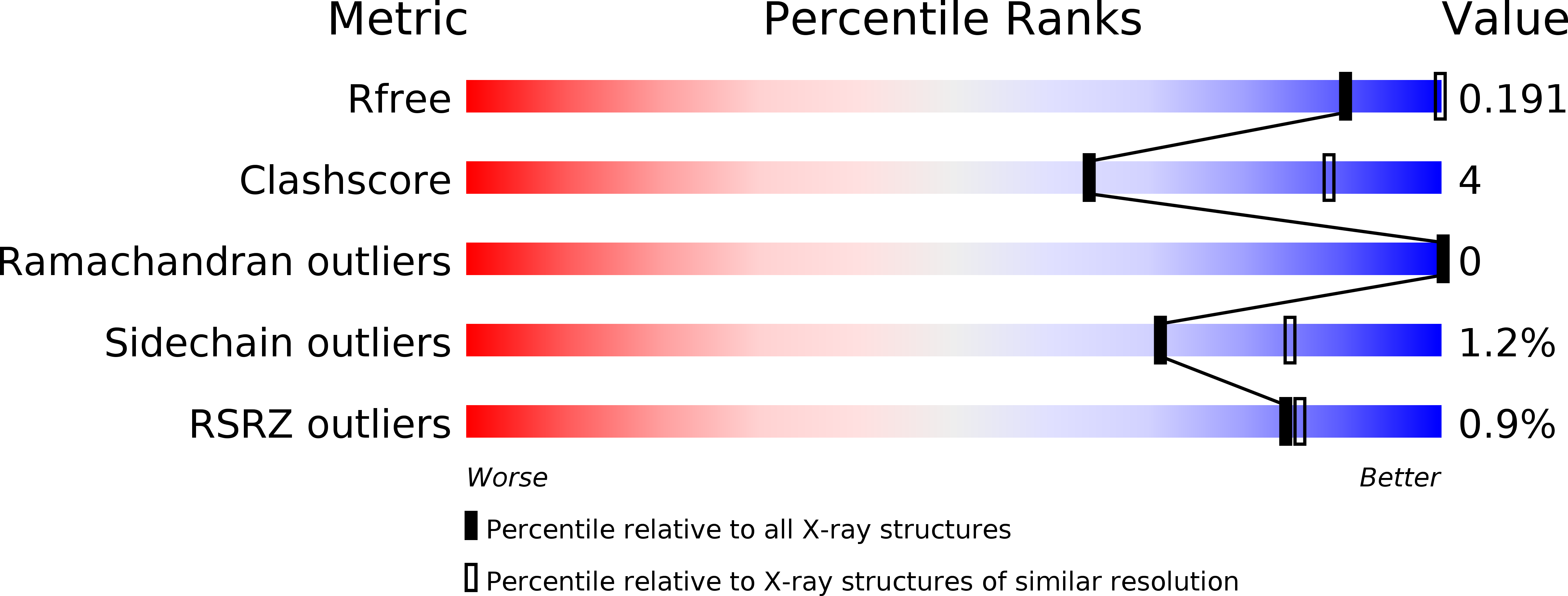
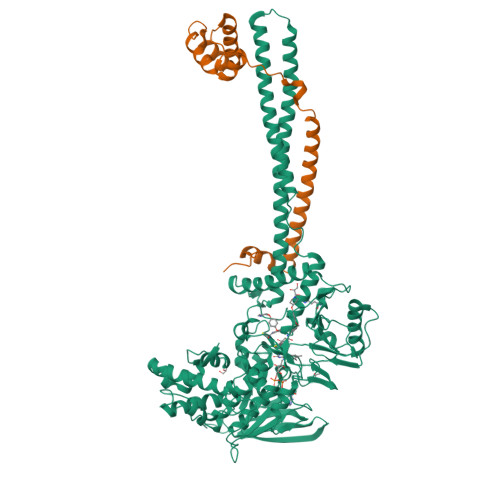
Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
Vianello, P., Sartori, L., Amigoni, F., Cappa, A., Faga, G., Fattori, R., Legnaghi, E., Ciossani, G., Mattevi, A., Meroni, G., Moretti, L., Cecatiello, V., Pasqualato, S., Romussi, A., Thaler, F., Trifiro, P., Villa, M., Botrugno, O.A., Dessanti, P., Minucci, S., Vultaggio, S., Zagarri, E., Varasi, M., Mercurio, C.(2017) J Med Chem 60: 1693-1715
- PubMed: 28186757
- DOI: https://doi.org/10.1021/acs.jmedchem.6b01019
- Primary Citation of Related Structures:
5LGT, 5LGU, 5LHG, 5LHH, 5LHI - PubMed Abstract:
The balance of methylation levels at histone H3 lysine 4 (H3K4) is regulated by KDM1A (LSD1). KDM1A is overexpressed in several tumor types, thus representing an emerging target for the development of novel cancer therapeutics. We have previously described ( Part 1, DOI 10.1021.acs.jmedchem.6b01018 ) the identification of thieno[3,2-b]pyrrole-5-carboxamides as novel reversible inhibitors of KDM1A, whose preliminary exploration resulted in compound 2 with biochemical IC 50 = 160 nM. We now report the structure-guided optimization of this chemical series based on multiple ligand/KDM1A-CoRest cocrystal structures, which led to several extremely potent inhibitors. In particular, compounds 46, 49, and 50 showed single-digit nanomolar IC 50 values for in vitro inhibition of KDM1A, with high selectivity in secondary assays. In THP-1 cells, these compounds transcriptionally affected the expression of genes regulated by KDM1A such as CD14, CD11b, and CD86. Moreover, 49 and 50 showed a remarkable anticlonogenic cell growth effect on MLL-AF9 human leukemia cells.
Organizational Affiliation:
Department of Experimental Oncology, Academic Drug Discovery, European Institute of Oncology , Via Adamello 16, 20139 Milano, Italy.