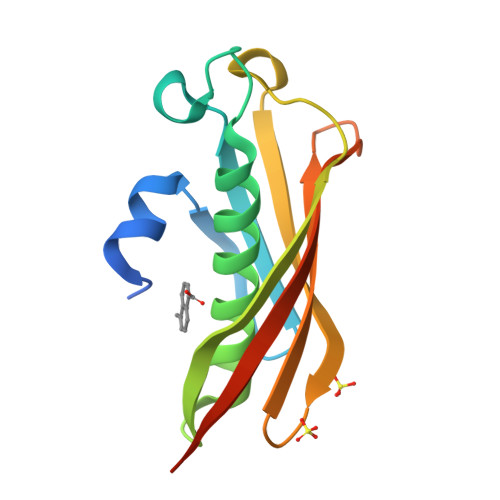
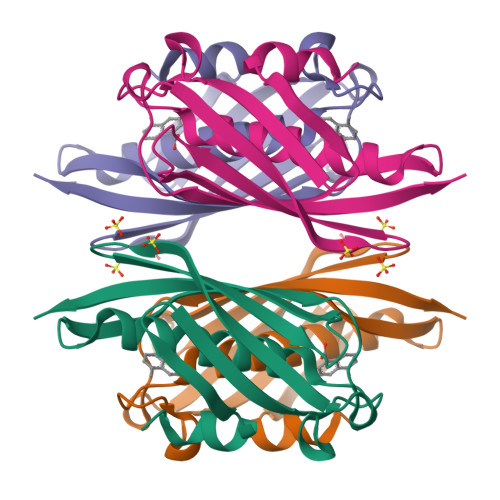
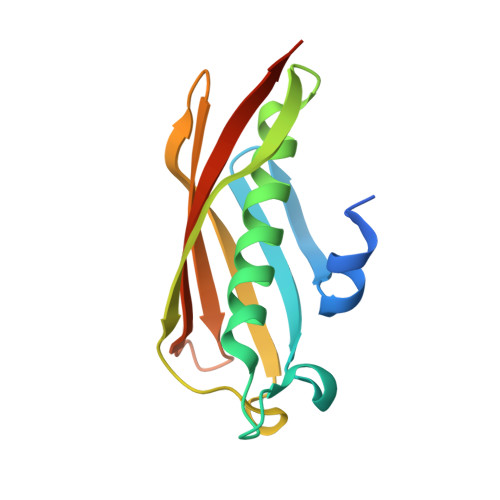
Polyketide Ring Expansion Mediated by a Thioesterase, Chain Elongation and Cyclization Domain, in Azinomycin Biosynthesis: Characterization of AziB and AziG.
Mori, S., Simkhada, D., Zhang, H., Erb, M.S., Zhang, Y., Williams, H., Fedoseyenko, D., Russell, W.K., Kim, D., Fleer, N., Ealick, S.E., Watanabe, C.M.(2016) Biochemistry 55: 704-714
- PubMed: 26731610
- DOI: https://doi.org/10.1021/acs.biochem.5b01050
- Primary Citation of Related Structures:
5HMB, 5HMC - PubMed Abstract:
The azinomycins are a family of potent antitumor agents with the ability to form interstrand cross-links with DNA. This study reports on the unusual biosynthetic formation of the 5-methyl naphthoate moiety, which is essential for effective DNA association. While sequence analysis predicts that the polyketide synthase (AziB) catalyzes the formation of this naphthoate, 2-methylbenzoic acid, a truncated single-ring product, is formed instead. We demonstrate that the thioesterase (AziG) acts as a chain elongation and cyclization (CEC) domain and is required for the additional two rounds of chain extension to form the expected product.
Organizational Affiliation:
Department of Chemistry, Texas A&M University , College Station, Texas 77843, United States.