Crystal structures of the selenoprotein glutathione peroxidase 4 in its apo form and in complex with the covalently bound inhibitor ML162.
Moosmayer, D., Hilpmann, A., Hoffmann, J., Schnirch, L., Zimmermann, K., Badock, V., Furst, L., Eaton, J.K., Viswanathan, V.S., Schreiber, S.L., Gradl, S., Hillig, R.C.(2021) Acta Crystallogr D Struct Biol 77: 237-248
- PubMed: 33559612
- DOI: https://doi.org/10.1107/S2059798320016125
- Primary Citation of Related Structures:
6HKQ, 6HN3 - PubMed Abstract:
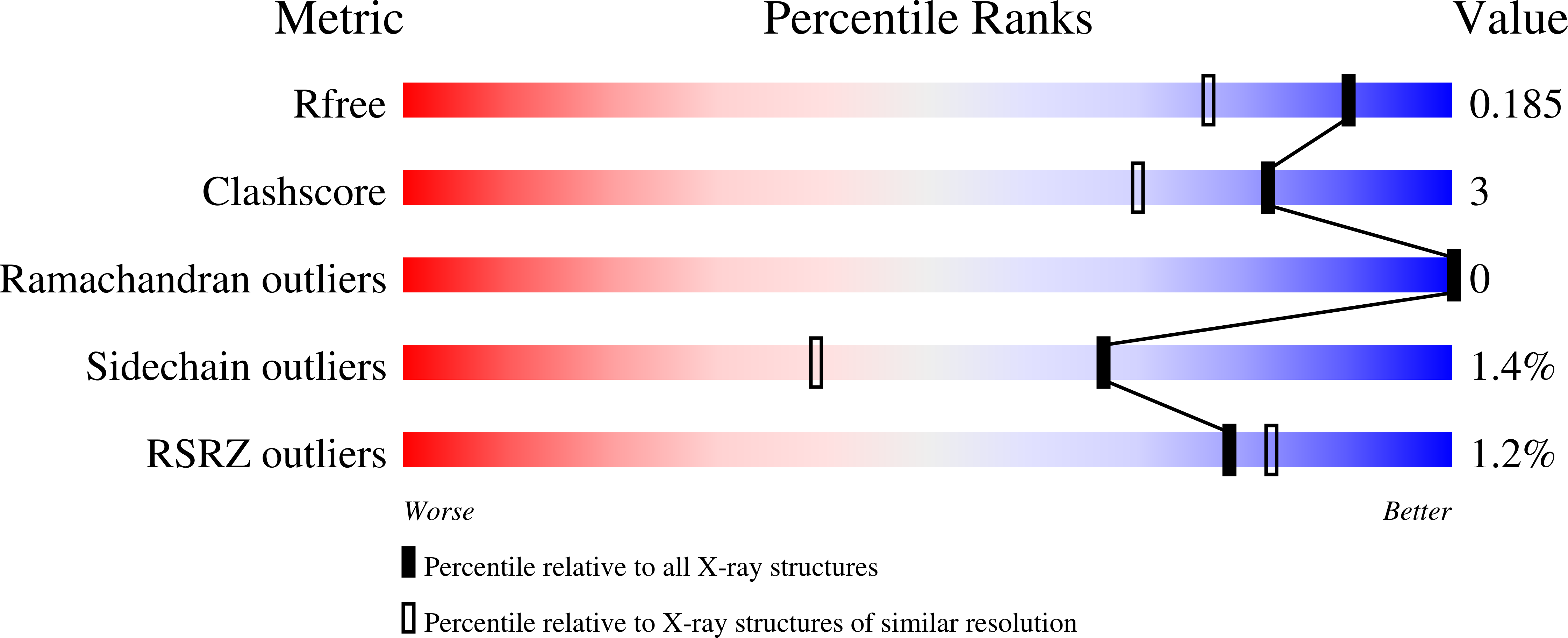
Wild-type human glutathione peroxidase 4 (GPX4) was co-expressed with SBP2 (selenocysteine insertion sequence-binding protein 2) in human HEK cells to achieve efficient production of this selenocysteine-containing enzyme on a preparative scale for structural biology. The protein was purified and crystallized, and the crystal structure of the wild-type form of GPX4 was determined at 1.0 Å resolution. The overall fold and the active site are conserved compared with previously determined crystal structures of mutated forms of GPX4. A mass-spectrometry-based approach was developed to monitor the reaction of the active-site selenocysteine Sec46 with covalent inhibitors. This, together with the introduction of a surface mutant (Cys66Ser), enabled the crystal structure determination of GPX4 in complex with the covalent inhibitor ML162 [(S)-enantiomer]. The mass-spectrometry-based approach described here opens the path to further co-complex crystal structures of this potential cancer drug target in complex with covalent inhibitors.
Organizational Affiliation:
Research and Development, Pharmaceuticals, Bayer AG, 13353 Berlin, Germany.