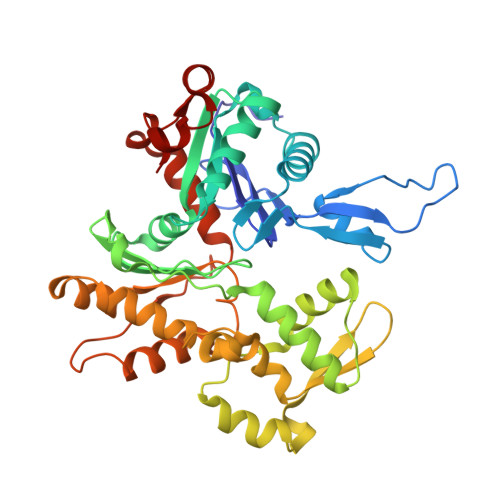
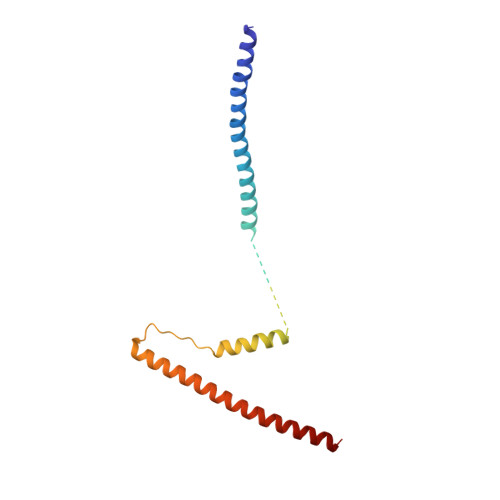
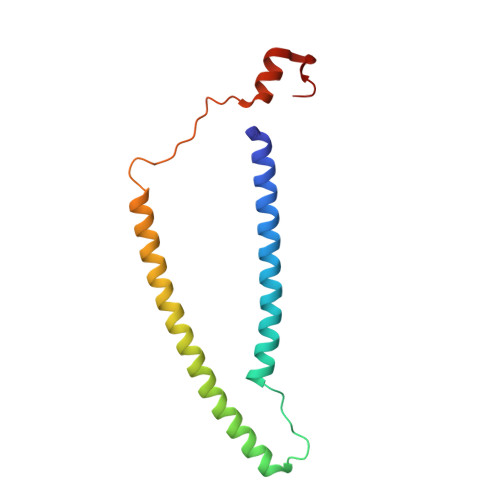
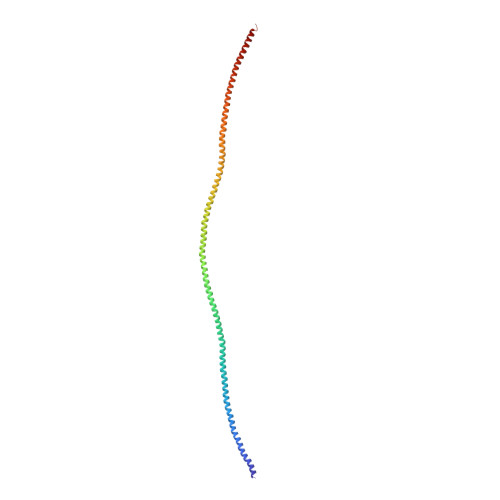Cardiac muscle thin filament structures reveal calcium regulatory mechanism.
Yamada, Y., Namba, K., Fujii, T.(2020) Nat Commun 11: 153-153
- PubMed: 31919429
- DOI: https://doi.org/10.1038/s41467-019-14008-1
- Primary Citation of Related Structures:
6KN7, 6KN8 - PubMed Abstract:
Contraction of striated muscles is driven by cyclic interactions of myosin head projecting from the thick filament with actin filament and is regulated by Ca 2+ released from sarcoplasmic reticulum. Muscle thin filament consists of actin, tropomyosin and troponin, and Ca 2+ binding to troponin triggers conformational changes of troponin and tropomyosin to allow actin-myosin interactions. However, the structural changes involved in this regulatory mechanism remain unknown. Here we report the structures of human cardiac muscle thin filament in the absence and presence of Ca 2+ by electron cryomicroscopy. Molecular models in the two states built based on available crystal structures reveal the structures of a C-terminal region of troponin I and an N-terminal region of troponin T in complex with the head-to-tail junction of tropomyosin together with the troponin core on actin filament. Structural changes of the thin filament upon Ca 2+ binding now reveal the mechanism of Ca 2+ regulation of muscle contraction.
Organizational Affiliation:
Graduate School of Frontier Biosciences, Osaka University, 1-3 Yamadaoka, Suita, Osaka, 565-0871, Japan.