Macrocyclic Peptides Uncover a Novel Binding Mode for Reversible Inhibitors of LSD1.
Yang, J., Talibov, V.O., Peintner, S., Rhee, C., Poongavanam, V., Geitmann, M., Sebastiano, M.R., Simon, B., Hennig, J., Dobritzsch, D., Danielson, U.H., Kihlberg, J.(2020) ACS Omega 5: 3979-3995
- PubMed: 32149225
- DOI: https://doi.org/10.1021/acsomega.9b03493
- Primary Citation of Related Structures:
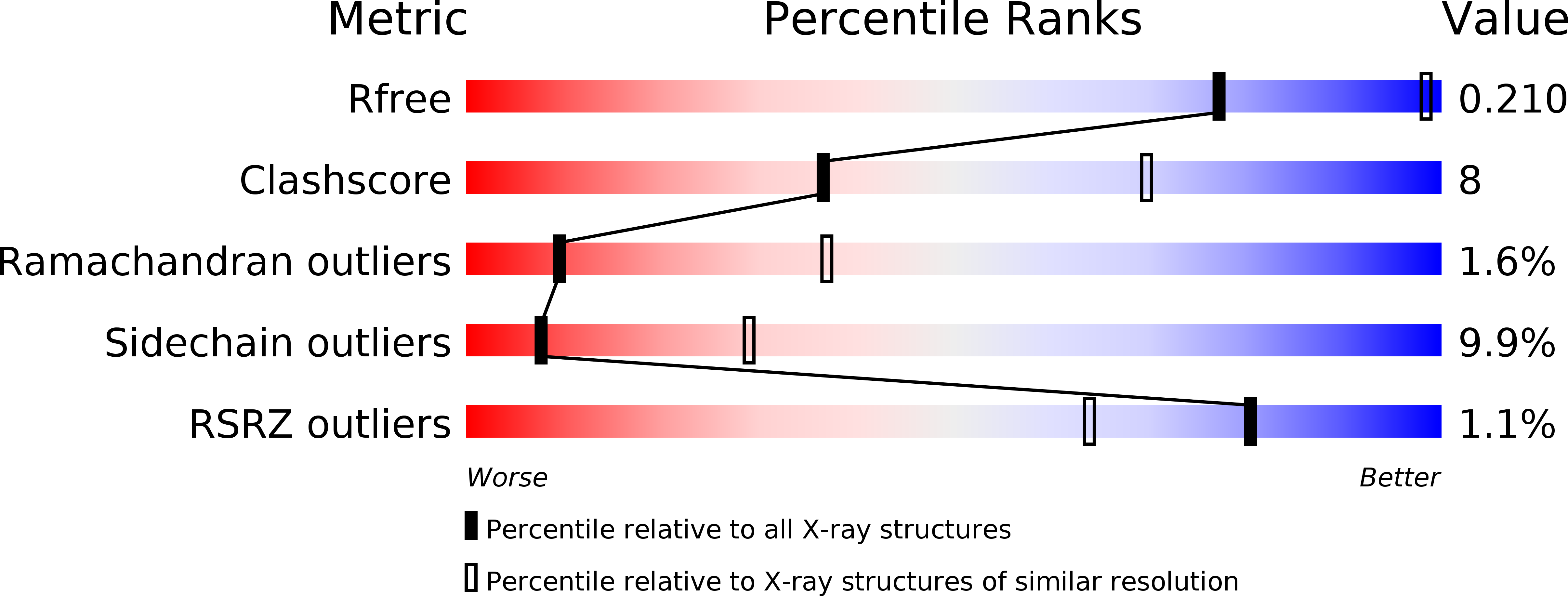
6S35 - PubMed Abstract:
Lysine-specific demethylase 1 (LSD1) is an epigenetic enzyme which regulates the methylation of Lys4 of histone 3 (H3) and is overexpressed in certain cancers. We used structures of H3 substrate analogues bound to LSD1 to design macrocyclic peptide inhibitors of LSD1. A linear, Lys4 to Met-substituted, 11-mer ( 4 ) was identified as the shortest peptide distinctly interacting with LSD1. It was evolved into macrocycle 31, which was >40 fold more potent ( K i = 2.3 μM) than 4 . Linear and macrocyclic peptides exhibited unexpected differences in structure-activity relationships for interactions with LSD1, indicating that they bind LSD1 differently. This was confirmed by the crystal structure of 31 in complex with LSD1-CoREST1, which revealed a novel binding mode at the outer rim of the LSD1 active site and without a direct interaction with FAD. NMR spectroscopy of 31 suggests that macrocyclization restricts its solution ensemble to conformations that include the one in the crystalline complex. Our results provide a solid basis for the design of optimized reversible LSD1 inhibitors.
Organizational Affiliation:
Department of Chemistry-BMC, Uppsala University, Box 576, SE-75123 Uppsala, Sweden.