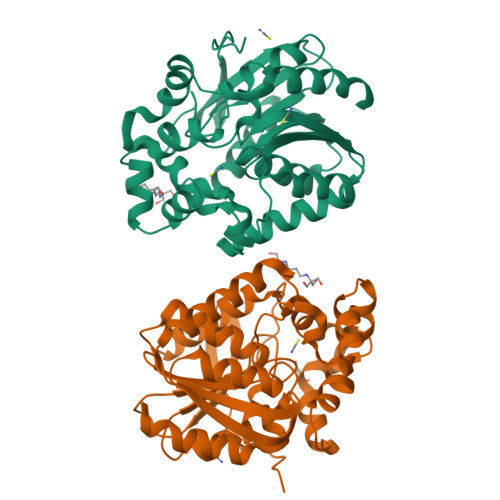
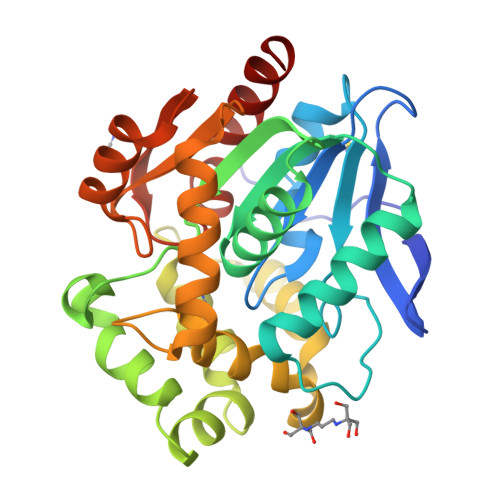
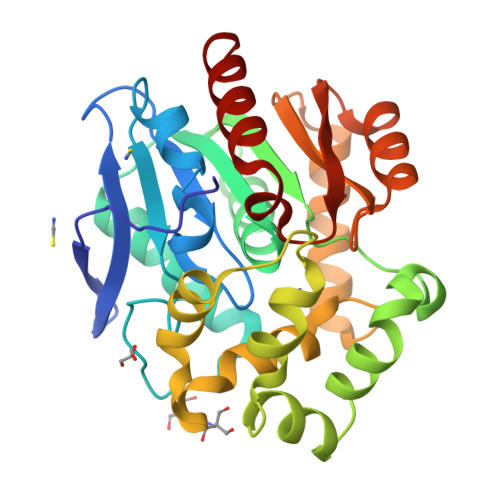
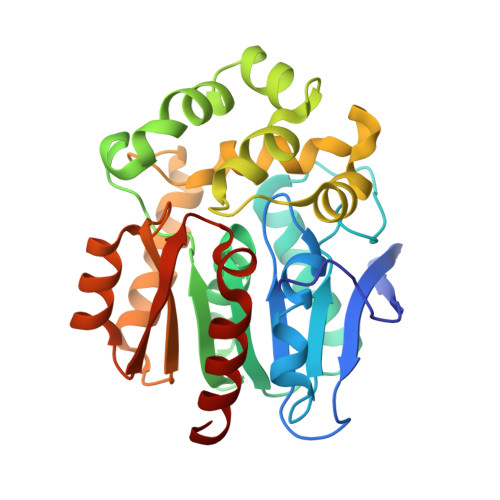
Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst.
Markova, K., Chmelova, K., Marques, S.M., Carpentier, P., Bednar, D., Damborsky, J., Marek, M.(2020) Chem Sci 11: 11162-11178
- PubMed: 34094357
- DOI: https://doi.org/10.1039/d0sc03367g
- Primary Citation of Related Structures:
6SP5, 6SP8 - PubMed Abstract:
Computational design of protein catalysts with enhanced stabilities for use in research and enzyme technologies is a challenging task. Using force-field calculations and phylogenetic analysis, we previously designed the haloalkane dehalogenase DhaA115 which contains 11 mutations that confer upon it outstanding thermostability ( T m = 73.5 °C; Δ T m > 23 °C). An understanding of the structural basis of this hyperstabilization is required in order to develop computer algorithms and predictive tools. Here, we report X-ray structures of DhaA115 at 1.55 Å and 1.6 Å resolutions and their molecular dynamics trajectories, which unravel the intricate network of interactions that reinforce the αβα-sandwich architecture. Unexpectedly, mutations toward bulky aromatic amino acids at the protein surface triggered long-distance (∼27 Å) backbone changes due to cooperative effects. These cooperative interactions produced an unprecedented double-lock system that: (i) induced backbone changes, (ii) closed the molecular gates to the active site, (iii) reduced the volumes of the main and slot access tunnels, and (iv) occluded the active site. Despite these spatial restrictions, experimental tracing of the access tunnels using krypton derivative crystals demonstrates that transport of ligands is still effective. Our findings highlight key thermostabilization effects and provide a structural basis for designing new thermostable protein catalysts.
Organizational Affiliation:
Loschmidt Laboratories, Department of Experimental Biology and RECETOX, Faculty of Science, Masaryk University Kamenice 5 625 00 Brno Czech Republic jiri@chemi.muni.cz martin.marek@recetox.muni.cz.