Structural insights into photoactivation and signalling in plant phytochromes.
Nagano, S., Guan, K., Shenkutie, S.M., Feiler, C., Weiss, M., Kraskov, A., Buhrke, D., Hildebrandt, P., Hughes, J.(2020) Nat Plants 6: 581-588
- PubMed: 32366982
- DOI: https://doi.org/10.1038/s41477-020-0638-y
- Primary Citation of Related Structures:
6TBY, 6TC5, 6TC7, 6TL4 - PubMed Abstract:
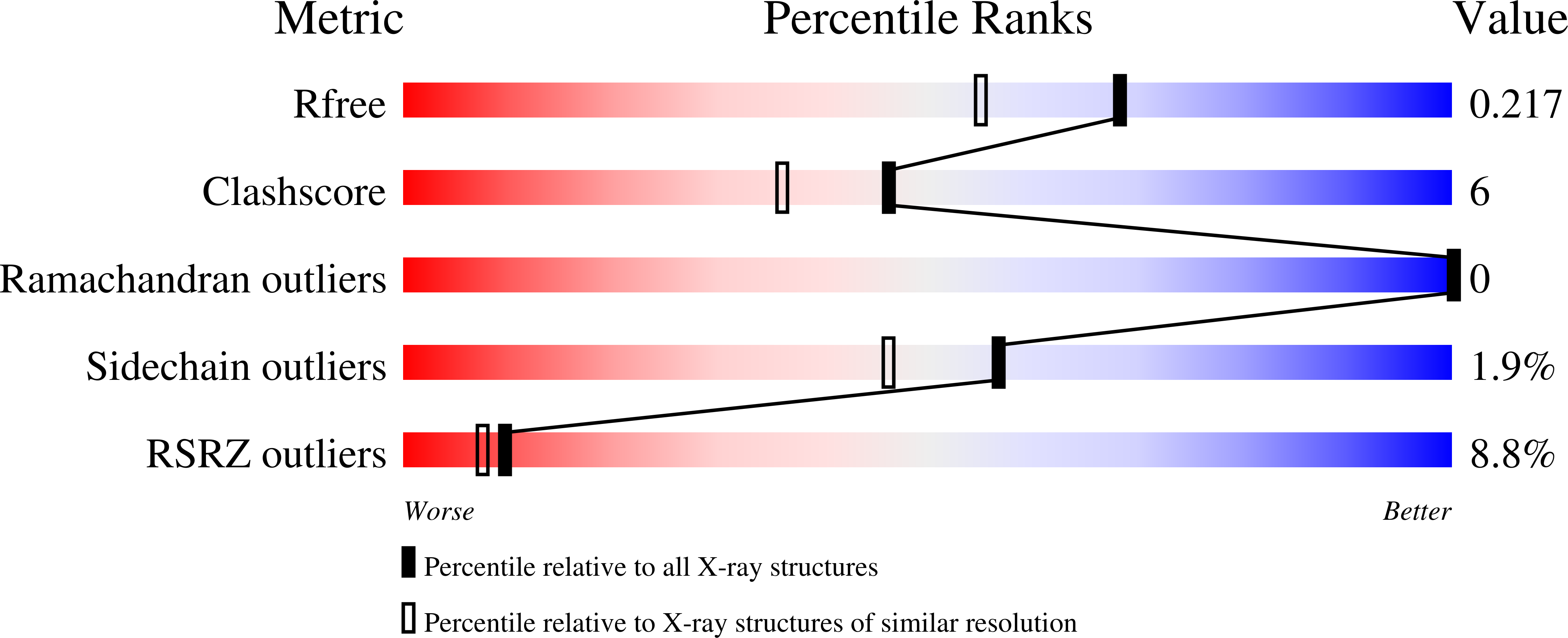
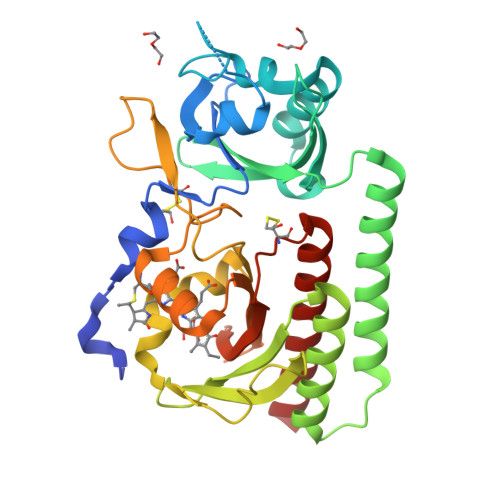
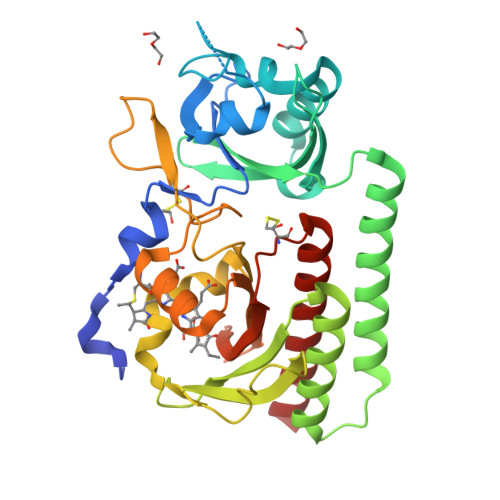
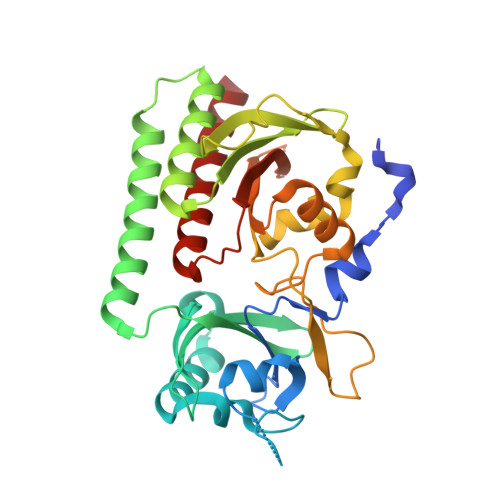
Plant phytochromes are red/far-red photochromic photoreceptors that act as master regulators of development, controlling the expression of thousands of genes. Here, we describe the crystal structures of four plant phytochrome sensory modules, three at about 2 Å resolution or better, including the first of an A-type phytochrome. Together with extensive spectral data, these structures provide detailed insight into the structure and function of plant phytochromes. In the Pr state, the substitution of phycocyanobilin and phytochromobilin cofactors has no structural effect, nor does the amino-terminal extension play a significant functional role. Our data suggest that the chromophore propionates and especially the phytochrome-specific domain tongue act differently in plant and prokaryotic phytochromes. We find that the photoproduct in period-ARNT-single-minded (PAS)-cGMP-specific phosphodiesterase-adenylyl cyclase-FhlA (GAF) bidomains might represent a novel intermediate between MetaRc and Pfr. We also discuss the possible role of a likely nuclear localization signal specific to and conserved in the phytochrome A lineage.
Organizational Affiliation:
Institut für Pflanzenphysiologie, Justus-Liebig-Universität, Gießen, Germany.