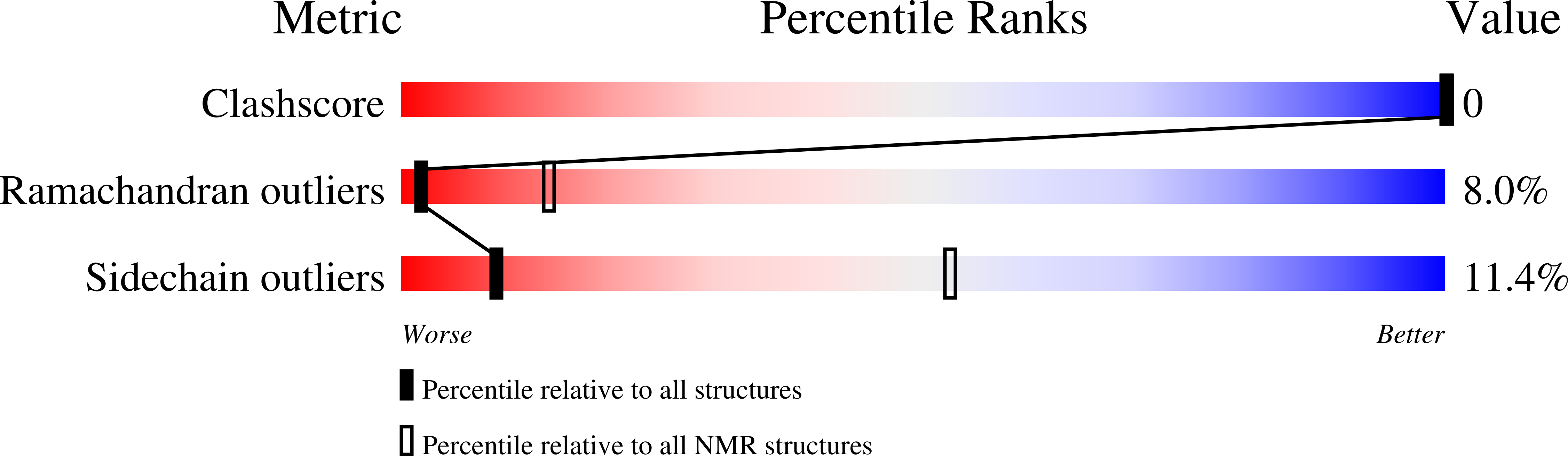Target-templated de novo design of macrocyclic d-/l-peptides: discovery of drug-like inhibitors of PD-1.
Guardiola, S., Varese, M., Roig, X., Sanchez-Navarro, M., Garcia, J., Giralt, E.(2021) Chem Sci 12: 5164-5170
- PubMed: 34163753
- DOI: https://doi.org/10.1039/d1sc01031j
- Primary Citation of Related Structures:
6TVJ - PubMed Abstract:
Peptides are a rapidly growing class of therapeutics with various advantages over traditional small molecules, especially for targeting difficult protein-protein interactions. However, current structure-based methods are largely limited to natural peptides and are not suitable for designing bioactive cyclic topologies that go beyond natural l-amino acids. Here, we report a generalizable framework that exploits the computational power of Rosetta, in terms of large-scale backbone sampling, side-chain composition and energy scoring, to design heterochiral cyclic peptides that bind to a protein surface of interest. To showcase the applicability of our approach, we developed two new inhibitors ( PD-i3 and PD-i6 ) of programmed cell death 1 (PD-1), a key immune checkpoint in oncology. A comprehensive biophysical evaluation was performed to assess their binding to PD-1 as well as their blocking effect on the endogenous PD-1/PD-L1 interaction. Finally, NMR elucidation of their in-solution structures confirmed our de novo design approach.
Organizational Affiliation:
Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology Baldiri Reixac 10 08028 Barcelona Spain ernest.giralt@irbbarcelona.org.