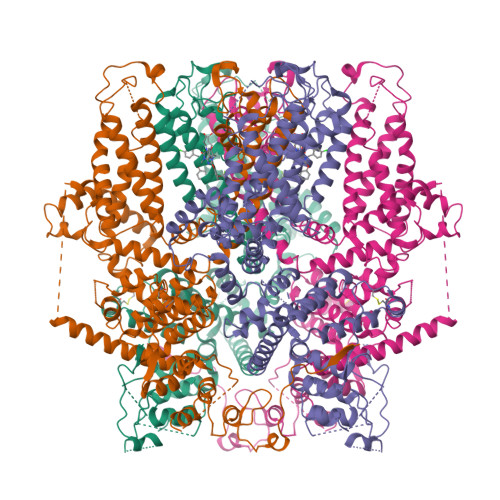
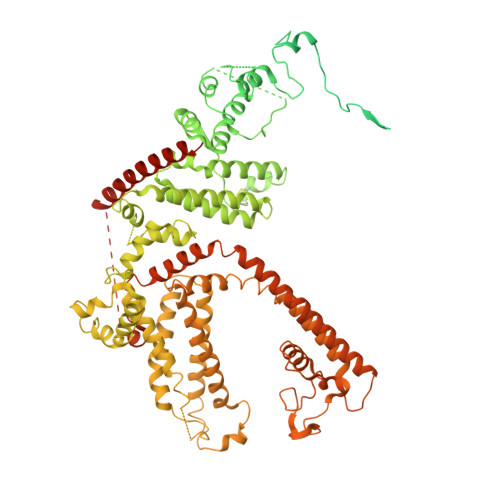
Human TRPC5 structures reveal interaction of a xanthine-based TRPC1/4/5 inhibitor with a conserved lipid binding site.
Wright, D.J., Simmons, K.J., Johnson, R.M., Beech, D.J., Muench, S.P., Bon, R.S.(2020) Commun Biol 3: 704-704
- PubMed: 33230284
- DOI: https://doi.org/10.1038/s42003-020-01437-8
- Primary Citation of Related Structures:
6YSN - PubMed Abstract:
TRPC1/4/5 channels are non-specific cation channels implicated in a wide variety of diseases, and TRPC1/4/5 inhibitors have recently entered clinical trials. However, fundamental and translational studies require a better understanding of TRPC1/4/5 channel regulation by endogenous and exogenous factors. Although several potent and selective TRPC1/4/5 modulators have been reported, the paucity of mechanistic insights into their modes-of-action remains a barrier to the development of new chemical probes and drug candidates. Xanthine-based modulators include the most potent and selective TRPC1/4/5 inhibitors described to date, as well as TRPC5 activators. Our previous studies suggest that xanthines interact with a, so far, elusive pocket of TRPC1/4/5 channels that is essential to channel gating. Here we report the structure of a small-molecule-bound TRPC1/4/5 channel-human TRPC5 in complex with the xanthine Pico145-to 3.0 Å. We found that Pico145 binds to a conserved lipid binding site of TRPC5, where it displaces a bound phospholipid. Our findings explain the mode-of-action of xanthine-based TRPC1/4/5 modulators, and suggest a structural basis for TRPC1/4/5 modulation by endogenous factors such as (phospho)lipids and Zn 2+ ions. These studies lay the foundations for the structure-based design of new generations of TRPC1/4/5 modulators.
Organizational Affiliation:
Discovery and Translational Science Department, Leeds Institute of Cardiovascular and Metabolic Medicine, University of Leeds, Leeds, LS2 9JT, UK.