Query-guided protein-protein interaction inhibitor discovery.
Celis, S., Hobor, F., James, T., Bartlett, G.J., Ibarra, A.A., Shoemark, D.K., Hegedus, Z., Hetherington, K., Woolfson, D.N., Sessions, R.B., Edwards, T.A., Andrews, D.M., Nelson, A., Wilson, A.J.(2021) Chem Sci 12: 4753-4762
- PubMed: 34163731
- DOI: https://doi.org/10.1039/d1sc00023c
- Primary Citation of Related Structures:
7A00 - PubMed Abstract:
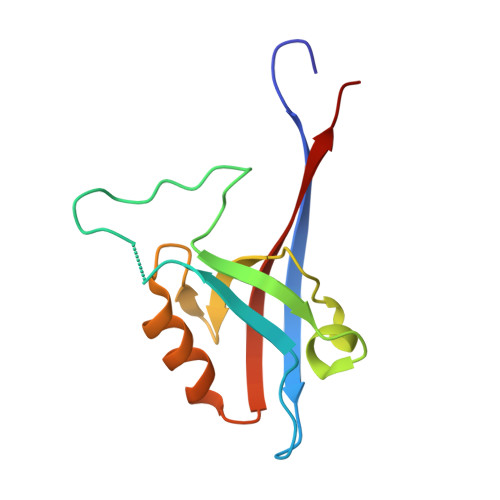
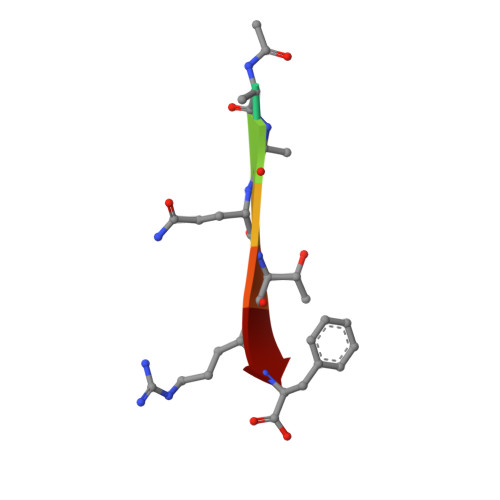
Protein-protein interactions (PPIs) are central to biological mechanisms, and can serve as compelling targets for drug discovery. Yet, the discovery of small molecule inhibitors of PPIs remains challenging given the large and typically shallow topography of the interacting protein surfaces. Here, we describe a general approach to the discovery of orthosteric PPI inhibitors that mimic specific secondary protein structures. Initially, hot residues at protein-protein interfaces are identified in silico or from experimental data, and incorporated into secondary structure-based queries. Virtual libraries of small molecules are then shape-matched against the queries, and promising ligands docked to target proteins. The approach is exemplified experimentally using two unrelated PPIs that are mediated by an α-helix (p53/ h DM2) and a β-strand (GKAP/SHANK1-PDZ). In each case, selective PPI inhibitors are discovered with low μM activity as determined by a combination of fluorescence anisotropy and 1 H- 15 N HSQC experiments. In addition, hit expansion yields a series of PPI inhibitors with defined structure-activity relationships. It is envisaged that the generality of the approach will enable discovery of inhibitors of a wide range of unrelated secondary structure-mediated PPIs.
Organizational Affiliation:
Astbury Centre for Structural Molecular Biology, University of Leeds Woodhouse Lane Leeds LS2 9JT UK a.j.wilson@leeds.ac.uk a.s.nelson@leeds.ac.uk.