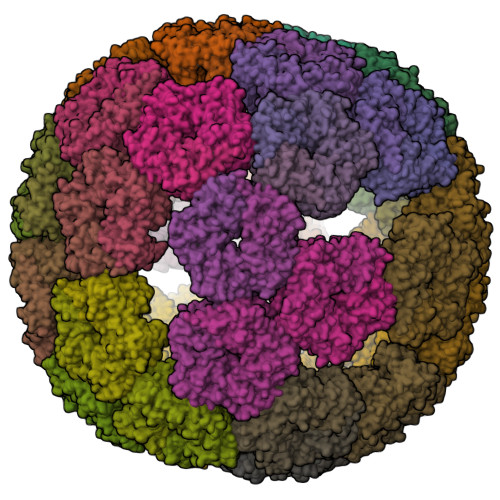
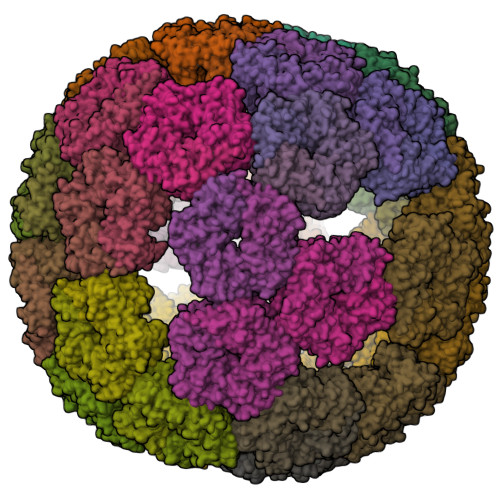
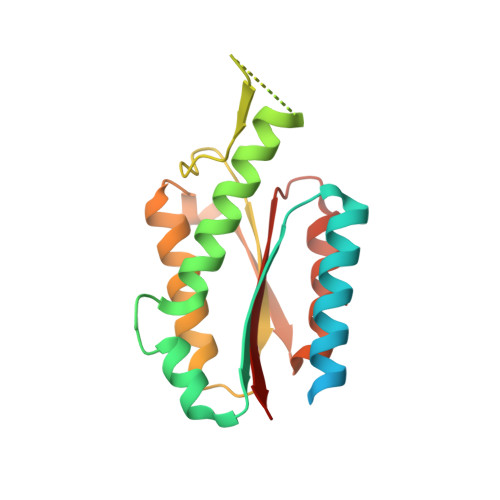
Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein.
Tetter, S., Terasaka, N., Steinauer, A., Bingham, R.J., Clark, S., Scott, A.J.P., Patel, N., Leibundgut, M., Wroblewski, E., Ban, N., Stockley, P.G., Twarock, R., Hilvert, D.(2021) Science 372: 1220-1224
- PubMed: 34112695
- DOI: https://doi.org/10.1126/science.abg2822
- Primary Citation of Related Structures:
7A4F, 7A4G, 7A4H, 7A4I, 7A4J - PubMed Abstract:
Viruses are ubiquitous pathogens of global impact. Prompted by the hypothesis that their earliest progenitors recruited host proteins for virion formation, we have used stringent laboratory evolution to convert a bacterial enzyme that lacks affinity for nucleic acids into an artificial nucleocapsid that efficiently packages and protects multiple copies of its own encoding messenger RNA. Revealing remarkable convergence on the molecular hallmarks of natural viruses, the accompanying changes reorganized the protein building blocks into an interlaced 240-subunit icosahedral capsid that is impermeable to nucleases, and emergence of a robust RNA stem-loop packaging cassette ensured high encapsidation yields and specificity. In addition to evincing a plausible evolutionary pathway for primordial viruses, these findings highlight practical strategies for developing nonviral carriers for diverse vaccine and delivery applications.
Organizational Affiliation:
Laboratory of Organic Chemistry, ETH Zurich, 8093 Zurich, Switzerland.