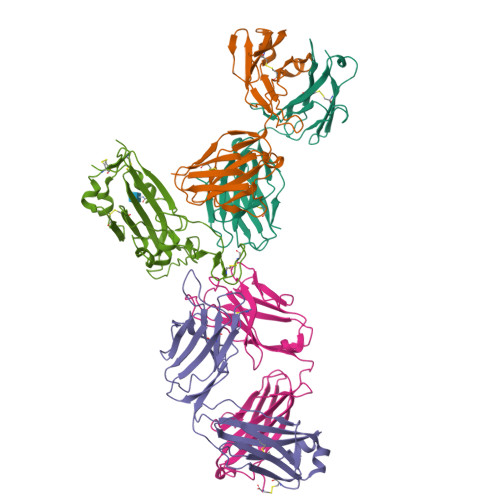
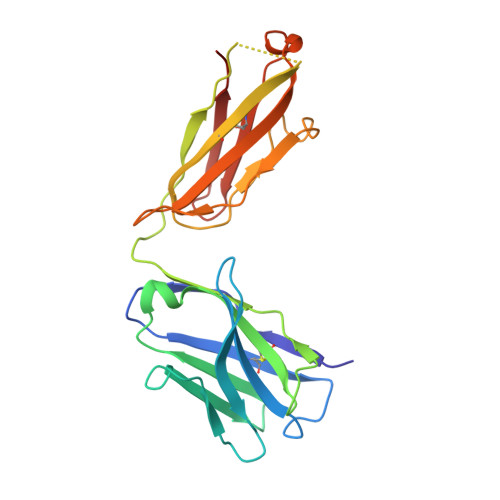
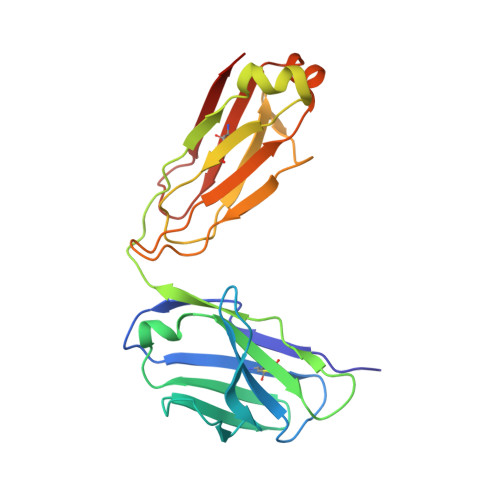
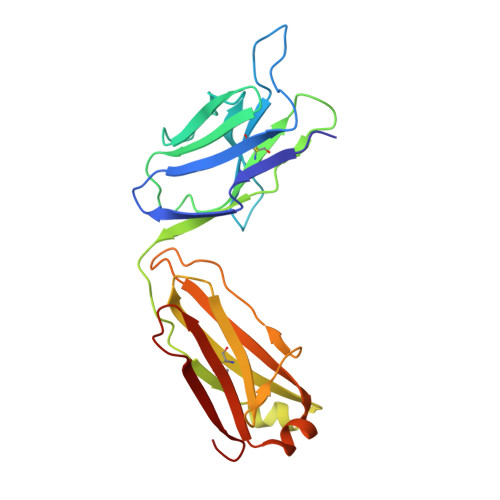
Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers.
Rujas, E., Kucharska, I., Tan, Y.Z., Benlekbir, S., Cui, H., Zhao, T., Wasney, G.A., Budylowski, P., Guvenc, F., Newton, J.C., Sicard, T., Semesi, A., Muthuraman, K., Nouanesengsy, A., Aschner, C.B., Prieto, K., Bueler, S.A., Youssef, S., Liao-Chan, S., Glanville, J., Christie-Holmes, N., Mubareka, S., Gray-Owen, S.D., Rubinstein, J.L., Treanor, B., Julien, J.P.(2021) Nat Commun 12: 3661-3661
- PubMed: 34135340
- DOI: https://doi.org/10.1038/s41467-021-23825-2
- Primary Citation of Related Structures:
7K9Z - PubMed Abstract:
SARS-CoV-2, the virus responsible for COVID-19, has caused a global pandemic. Antibodies can be powerful biotherapeutics to fight viral infections. Here, we use the human apoferritin protomer as a modular subunit to drive oligomerization of antibody fragments and transform antibodies targeting SARS-CoV-2 into exceptionally potent neutralizers. Using this platform, half-maximal inhibitory concentration (IC 50 ) values as low as 9 × 10 - 14 M are achieved as a result of up to 10,000-fold potency enhancements compared to corresponding IgGs. Combination of three different antibody specificities and the fragment crystallizable (Fc) domain on a single multivalent molecule conferred the ability to overcome viral sequence variability together with outstanding potency and IgG-like bioavailability. The MULTi-specific, multi-Affinity antiBODY (Multabody or MB) platform thus uniquely leverages binding avidity together with multi-specificity to deliver ultrapotent and broad neutralizers against SARS-CoV-2. The modularity of the platform also makes it relevant for rapid evaluation against other infectious diseases of global health importance. Neutralizing antibodies are a promising therapeutic for SARS-CoV-2.
Organizational Affiliation:
Program in Molecular Medicine, The Hospital for Sick Children Research Institute, Toronto, ON, Canada.