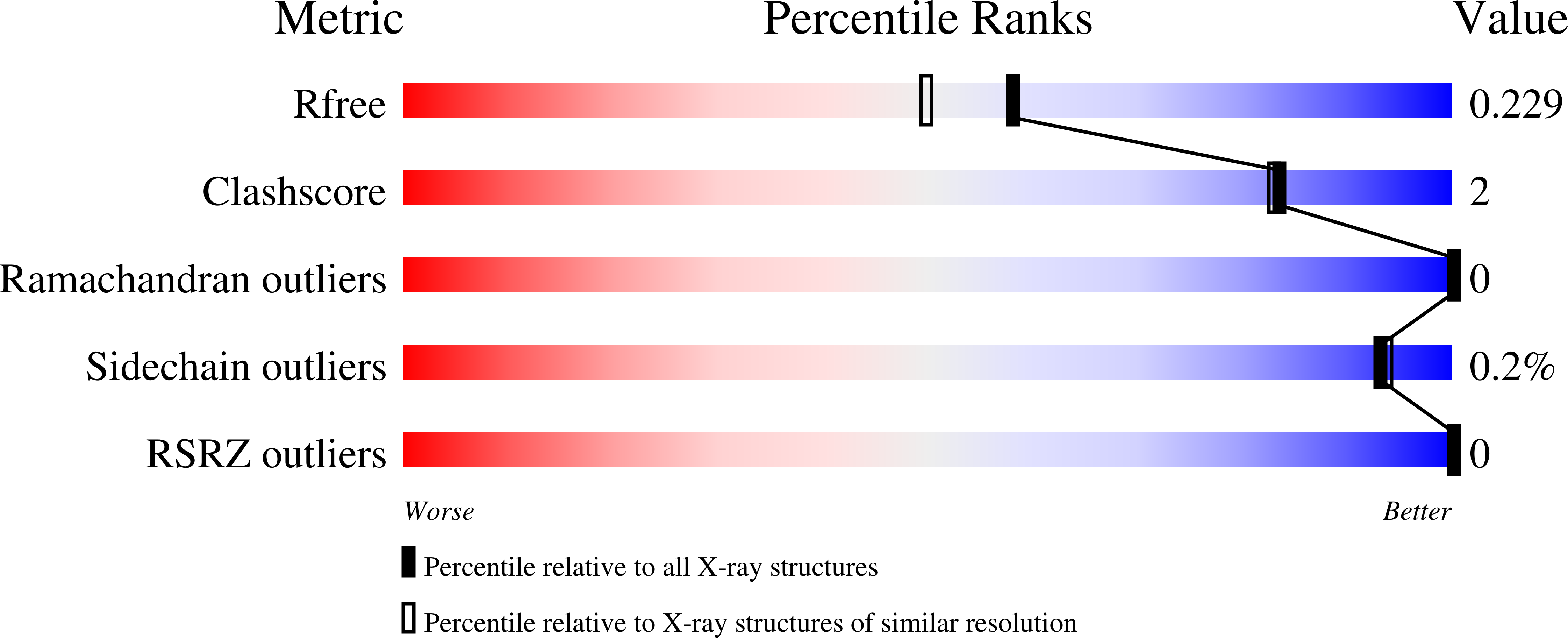
Machine-Directed Evolution of an Imine Reductase for Activity and Stereoselectivity
Ma, E.J., Siirola, E., Moore, C., Kummer, A., Stoeckli, M., Faller, M., Bouquet, C., Eggimann, F., Ligibel, M., Huynh, D., Cutler, G., Siegrist, L., Lewis, R.A., Acker, A.C., Freund, E., Koch, E., Vogel, M., Schlingensiepen, H., Oakeley, E.J., Snajdrova, R.(2021) ACS Catal : 12433-12445