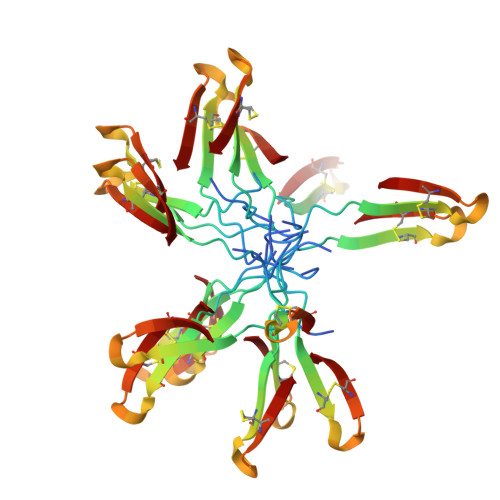
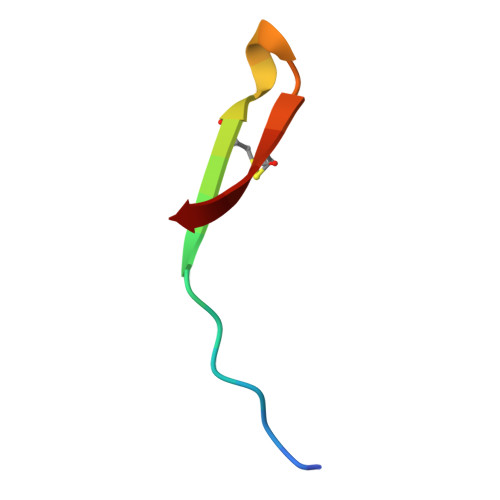
Solution structure of thanatin, a potent bactericidal and fungicidal insect peptide, determined from proton two-dimensional nuclear magnetic resonance data.
Mandard, N., Sodano, P., Labbe, H., Bonmatin, J.M., Bulet, P., Hetru, C., Ptak, M., Vovelle, F.(1998) Eur J Biochem 256: 404-410
- PubMed: 9760181
- DOI: https://doi.org/10.1046/j.1432-1327.1998.2560404.x
- Primary Citation of Related Structures:
8TFV - PubMed Abstract:
Thanatin is the first inducible insect peptide that has been found to have, at physiological concentrations, a broad range of activity against bacteria and fungi. Thanatin contains 21 amino acids including two cysteine residues that form a disulfide bridge. Two-dimensional (2D) 1H-NMR spectroscopy and molecular modelling have been used to determine its three-dimensional (3D) structure in water. Thanatin adopts a well-defined anti-parallel beta-sheet structure from residue 8 to the C-terminus, including the disulfide bridge. In spite of the presence of two proline residues, there is a large degree of structural variability in the N-terminal segment. The structure of thanatin is quite different from the known structures of other insect defence peptides, such as antibacterial defensin and antifungal drosomycin. It has more similarities with the structures of various peptides from different origins, such as brevinins, protegrins and tachyplesins, which have a two-stranded beta-sheet stabilized by one or two disulfide bridges. Combined with activity test experiments on several truncated isoforms of thanatin, carried out by Fehlbaum et al. [Fehlbaum, P., Bulet, P., Chernysh, S., Briand, J. P., Roussel, J. P., Letellier, L., Hétru, C. & Hoffmann, J. (1996) Proc. Natl Acad. Sci. USA 93, 1221-1225], our structural study evidences the importance of the beta-sheet structure and also suggests that anti-Gram-negative activity involves a site formed by the Arg20 side-chain embedded in a hydrophobic cluster.
Organizational Affiliation:
Centre de Biophysique Moléculaire, CNRS-UPR 4301, University of Orléans, France.